1YUB
 
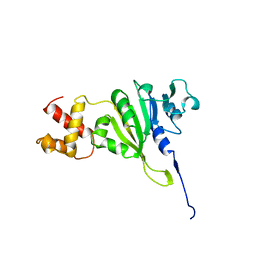 | | SOLUTION STRUCTURE OF AN RRNA METHYLTRANSFERASE (ERMAM) THAT CONFERS MACROLIDE-LINCOSAMIDE-STREPTOGRAMIN ANTIBIOTIC RESISTANCE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | RRNA METHYLTRANSFERASE | | Authors: | Yu, L, Petros, A.M, Schnuchel, A, Zhong, P, Severin, J.M, Walter, K, Holzman, T.F, Fesik, S.W. | | Deposit date: | 1997-03-04 | | Release date: | 1998-03-04 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an rRNA methyltransferase (ErmAM) that confers macrolide-lincosamide-streptogramin antibiotic resistance.
Nat.Struct.Biol., 4, 1997
|
|
1DHM
 
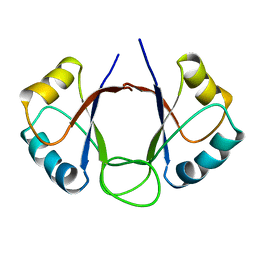 | | DNA-BINDING DOMAIN OF E2 FROM HUMAN PAPILLOMAVIRUS-31, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | E2 PROTEIN | | Authors: | Liang, H, Petros, A.P, Meadows, R.P, Yoon, H.S, Egan, D.A, Walter, K, Holzman, T.F, Robins, T, Fesik, S.W. | | Deposit date: | 1995-08-15 | | Release date: | 1996-12-07 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of a human papillomavirus E2 protein: evidence for flexible DNA-binding regions.
Biochemistry, 35, 1996
|
|
1A7G
 
 | |
2ERC
 
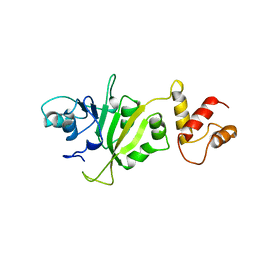 | | CRYSTAL STRUCTURE OF ERMC' A RRNA-METHYL TRANSFERASE | | Descriptor: | RRNA METHYL TRANSFERASE | | Authors: | Bussiere, D.E, Muchmore, S.W, Dealwis, C.G, Schluckebier, G, Abad-Zapatero, C. | | Deposit date: | 1998-03-13 | | Release date: | 1999-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Crystal structure of ErmC', an rRNA methyltransferase which mediates antibiotic resistance in bacteria.
Biochemistry, 37, 1998
|
|
1CYB
 
 | |
1CYA
 
 | |
2AM1
 
 | | sp protein ligand 1 | | Descriptor: | 2,4-DICHLORO-N-(3-CYANO-4,5,6,7-TETRAHYDRO-BENZOTHIOPHEN-2YL)-5-(MORPHOLINE-4-SULFONYL)-BENZAMIDE, GLYCEROL, UDP-N-acetylmuramoylalanine-D-glutamyl-lysine-D-alanyl-D-alanine ligase, ... | | Authors: | Longenecker, K.L, Stamper, G.F, Hajduk, P.J, Fry, E.H, Jakob, C.G, Harlan, J.E, Edalji, R, Bartley, D.M, Walter, K.A, Solomon, L.R. | | Deposit date: | 2005-08-08 | | Release date: | 2006-01-24 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of MurF from Streptococcus pneumoniae co-crystallized with a small molecule inhibitor exhibits interdomain closure
Protein Sci., 14, 2005
|
|
2AM2
 
 | | sp protein ligand 2 | | Descriptor: | 2-CHLORO-N-(3-CYANO-5,6-DIHYDRO-4H-CYCLOPENTA[B]THIOPHEN-2-YL)-5-DIETHYLSULFAMOYL-BENZAMIDE, UDP-N-acetylmuramoylalanine-D-glutamyl-lysine-D-alanyl-D-alanine ligase, MurF protein | | Authors: | Longenecker, K.L, Stamper, G.F, Hajduk, P.J, Fry, E.H, Jakob, C.G, Harlan, J.E, Edalji, R, Bartley, D.M, Walter, K.A, Solomon, L.R. | | Deposit date: | 2005-08-08 | | Release date: | 2006-01-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of MurF from Streptococcus pneumoniae co-crystallized with a small molecule inhibitor exhibits interdomain closure
Protein Sci., 14, 2005
|
|
1ZP5
 
 | | Crystal structure of the complex between MMP-8 and a N-hydroxyurea inhibitor | | Descriptor: | CALCIUM ION, N-{2-[(4'-CYANO-1,1'-BIPHENYL-4-YL)OXY]ETHYL}-N'-HYDROXY-N-METHYLUREA, Neutrophil collagenase, ... | | Authors: | Campestre, C, Agamennone, M, Tortorella, P, Preziuso, S, Biasone, A, Gavuzzo, E, Pochetti, G, Mazza, F, Tschesche, H, Gallina, C. | | Deposit date: | 2005-05-16 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | N-Hydroxyurea as zinc binding group in matrix metalloproteinase inhibition: Mode of binding in a complex with MMP-8.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1QAN
 
 | | THE STRUCTURE OF THE RRNA METHYLTRANSFERASE ERMC': IMPLICATIONS FOR THE REACTION MECHANISM | | Descriptor: | ACETATE ION, ERMC' METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Schluckebier, G, Zhong, P, Stewart, K.D, Kavanaugh, T.J, Abad-Zapatero, C. | | Deposit date: | 1999-03-26 | | Release date: | 2000-03-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The 2.2 A structure of the rRNA methyltransferase ErmC' and its complexes with cofactor and cofactor analogs: implications for the reaction mechanism.
J.Mol.Biol., 289, 1999
|
|
1QAO
 
 | | THE STRUCTURE OF THE RRNA METHYLTRANSFERASE ERMC': IMPLICATIONS FOR THE REACTION MECHANISM | | Descriptor: | ERMC' METHYLTRANSFERASE, S-ADENOSYLMETHIONINE | | Authors: | Schluckebier, G, Zhong, P, Stewart, K.D, Kavanaugh, T.J, Abad-Zapatero, C. | | Deposit date: | 1999-03-26 | | Release date: | 2000-03-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The 2.2 A structure of the rRNA methyltransferase ErmC' and its complexes with cofactor and cofactor analogs: implications for the reaction mechanism.
J.Mol.Biol., 289, 1999
|
|
1QAM
 
 | | THE STRUCTURE OF THE RRNA METHYLTRANSFERASE ERMC': IMPLICATIONS FOR THE REACTION MECHANISM | | Descriptor: | ACETATE ION, ERMC' METHYLTRANSFERASE | | Authors: | Schluckebier, G, Zhong, P, Stewart, K.D, Kavanaugh, T.J, Abad-Zapatero, C. | | Deposit date: | 1999-03-25 | | Release date: | 2000-03-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 2.2 A structure of the rRNA methyltransferase ErmC' and its complexes with cofactor and cofactor analogs: implications for the reaction mechanism.
J.Mol.Biol., 289, 1999
|
|
1QAQ
 
 | | THE STRUCTURE OF THE RRNA METHYLTRANSFERASE ERMC': IMPLICATIONS FOR THE REACTION MECHANISM | | Descriptor: | ERMC' RRNA METHYLTRANSFERASE, SINEFUNGIN | | Authors: | Schluckebier, G, Zhong, P, Stewart, K.D, Kavanaugh, T.J, Abad-Zapatero, C. | | Deposit date: | 1999-03-28 | | Release date: | 2000-03-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The 2.2 A structure of the rRNA methyltransferase ErmC' and its complexes with cofactor and cofactor analogs: implications for the reaction mechanism.
J.Mol.Biol., 289, 1999
|
|
