2IEW
 
 | |
2IF8
 
 | |
6YSR
 
 | | Structure of the P+9 stalled ribosome complex | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Chan, K.-H, Petrychenko, V, Mueller, C, Maracci, C, Holtkamp, W, Wilson, D.N, Fischer, N, Rodnina, M.V. | | Deposit date: | 2020-04-23 | | Release date: | 2020-08-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of ribosome rescue by alternative ribosome-rescue factor B.
Nat Commun, 11, 2020
|
|
6YSU
 
 | | Structure of the P+0 ArfB-ribosome complex in the post-hydrolysis state | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Chan, K.-H, Petrychenko, V, Mueller, C, Maracci, C, Holtkamp, W, Wilson, D.N, Fischer, N, Rodnina, M.V. | | Deposit date: | 2020-04-23 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanism of ribosome rescue by alternative ribosome-rescue factor B.
Nat Commun, 11, 2020
|
|
6YSS
 
 | | Structure of the P+9 ArfB-ribosome complex in the post-hydrolysis state | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Chan, K.-H, Petrychenko, V, Mueller, C, Maracci, C, Holtkamp, W, Wilson, D.N, Fischer, N, Rodnina, M.V. | | Deposit date: | 2020-04-23 | | Release date: | 2020-08-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Mechanism of ribosome rescue by alternative ribosome-rescue factor B.
Nat Commun, 11, 2020
|
|
6YST
 
 | | Structure of the P+9 ArfB-ribosome complex with P/E hybrid tRNA in the post-hydrolysis state | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Chan, K.-H, Petrychenko, V, Mueller, C, Maracci, C, Holtkamp, W, Wilson, D.N, Fischer, N, Rodnina, M.V. | | Deposit date: | 2020-04-23 | | Release date: | 2020-08-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of ribosome rescue by alternative ribosome-rescue factor B.
Nat Commun, 11, 2020
|
|
1C25
 
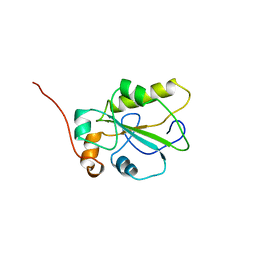 | | HUMAN CDC25A CATALYTIC DOMAIN | | Descriptor: | CDC25A | | Authors: | Fauman, E.B, Cogswell, J.P, Lovejoy, B, Rocque, W.J, Holmes, W, Montana, V.G, Piwnica-Worms, H, Rink, M.J, Saper, M.A. | | Deposit date: | 1998-04-17 | | Release date: | 1998-08-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the catalytic domain of the human cell cycle control phosphatase, Cdc25A.
Cell(Cambridge,Mass.), 93, 1998
|
|
4NSY
 
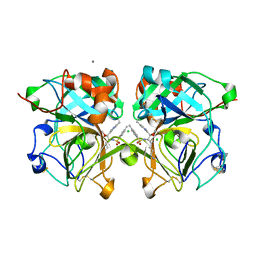 | | Wild-type lysobacter enzymogenes lysc endoproteinase covalently inhibited by TLCK | | Descriptor: | CALCIUM ION, CHLORIDE ION, Lysyl endopeptidase, ... | | Authors: | Asztalos, P, Muller, A, Holke, W, Sobek, H, Rudolph, M.G. | | Deposit date: | 2013-11-29 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic resolution structure of a lysine-specific endoproteinase from Lysobacter enzymogenes suggests a hydroxyl group bound to the oxyanion hole.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NSV
 
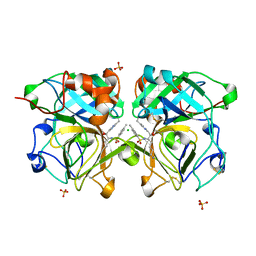 | | Lysobacter enzymogenes lysc endoproteinase K30R mutant covalently inhibited by TLCK | | Descriptor: | CHLORIDE ION, Lysyl endopeptidase, N-[(2S,3S)-7-amino-1-chloro-2-hydroxyheptan-3-yl]-4-methylbenzenesulfonamide (Bound Form), ... | | Authors: | Asztalos, P, Muller, A, Holke, W, Sobek, H, Rudolph, M.G. | | Deposit date: | 2013-11-29 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Atomic resolution structure of a lysine-specific endoproteinase from Lysobacter enzymogenes suggests a hydroxyl group bound to the oxyanion hole.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3K81
 
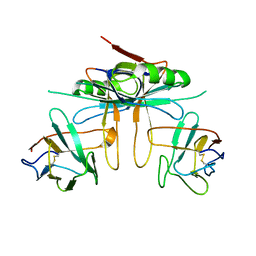 | |
3STB
 
 | | A complex of two editosome proteins and two nanobodies | | Descriptor: | MP18 RNA editing complex protein, RNA-editing complex protein MP42, single domain antibody VHH | | Authors: | Park, Y.-J, Hol, W. | | Deposit date: | 2011-07-09 | | Release date: | 2011-11-02 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a heterodimer of editosome interaction proteins in complex with two copies of a cross-reacting nanobody.
Nucleic Acids Res., 40, 2012
|
|
4DK3
 
 | | Structure of Editosome protein | | Descriptor: | RNA-editing complex protein MP81, single domain antibody VHH | | Authors: | Park, Y.-J, Hol, W. | | Deposit date: | 2012-02-03 | | Release date: | 2012-07-04 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | The structure of the C-terminal domain of the largest editosome interaction protein and its role in promoting RNA binding by RNA-editing ligase L2.
Nucleic Acids Res., 40, 2012
|
|
4DKA
 
 | | Structure of Editosome protein | | Descriptor: | RNA-editing complex protein MP81, SODIUM ION, single domain antibody VHH | | Authors: | Park, Y.-J, Hol, W. | | Deposit date: | 2012-02-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The structure of the C-terminal domain of the largest editosome interaction protein and its role in promoting RNA binding by RNA-editing ligase L2.
Nucleic Acids Res., 40, 2012
|
|
4DNI
 
 | | Structure of Editosome protein | | Descriptor: | Fusion protein of RNA-editing complex proteins MP42 and MP18 | | Authors: | Park, Y.-J, Hol, W. | | Deposit date: | 2012-02-08 | | Release date: | 2012-12-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Explorations of linked editosome domains leading to the discovery of motifs defining conserved pockets in editosome OB-folds.
J.Struct.Biol., 180, 2012
|
|
4DK6
 
 | | Structure of Editosome protein | | Descriptor: | RNA-editing complex protein MP81, single domain antibody VHH | | Authors: | Park, Y.-J, Hol, W. | | Deposit date: | 2012-02-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The structure of the C-terminal domain of the largest editosome interaction protein and its role in promoting RNA binding by RNA-editing ligase L2.
Nucleic Acids Res., 40, 2012
|
|
4PHT
 
 | | ATPase GspE in complex with the cytoplasmic domain of GspL from the Vibrio vulnificus type II Secretion system | | Descriptor: | General secretory pathway protein E, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Lu, C, Korotkov, K, Hol, W. | | Deposit date: | 2014-05-06 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Crystal structure of the full-length ATPase GspE from the Vibrio vulnificus type II secretion system in complex with the cytoplasmic domain of GspL.
J.Struct.Biol., 187, 2014
|
|
1MC0
 
 | | Regulatory Segment of Mouse 3',5'-Cyclic Nucleotide Phosphodiesterase 2A, Containing the GAF A and GAF B Domains | | Descriptor: | 3',5'-cyclic nucleotide phosphodiesterase 2A, CYCLIC GUANOSINE MONOPHOSPHATE | | Authors: | Martinez, S, Wu, A, Glavas, N, Tang, X, Turley, S, Hol, W, Beavo, J. | | Deposit date: | 2002-08-04 | | Release date: | 2002-10-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | The two GAF domains in phosphodiesterase 2A have distinct roles in dimerization and in cGMP binding.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1SKZ
 
 | | PROTEASE INHIBITOR | | Descriptor: | ANTISTASIN, CHLORIDE ION | | Authors: | Krengel, U, Dijkstra, B.W. | | Deposit date: | 1997-04-16 | | Release date: | 1997-10-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of antistasin at 1.9 A resolution and its modelled complex with blood coagulation factor Xa.
EMBO J., 16, 1997
|
|
1HTL
 
 | |
1LT4
 
 | | HEAT-LABILE ENTEROTOXIN MUTANT S63K | | Descriptor: | HEAT-LABILE ENTEROTOXIN, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Van Den Akker, F, Hol, W.G.J. | | Deposit date: | 1997-04-14 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a non-toxic mutant of heat-labile enterotoxin, which is a potent mucosal adjuvant.
Protein Sci., 6, 1997
|
|
1LTG
 
 | |
1LPF
 
 | |
1GYQ
 
 | |
6QBA
 
 | | Crystal Structure of Retinol-Binding Protein 4 (RBP4) in complex with non-retinoid ligand A1120 and engineered binding scaffold | | Descriptor: | 2-[({4-[2-(trifluoromethyl)phenyl]piperidin-1-yl}carbonyl)amino]benzoic acid, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mlynek, G, Brey, C.U, Djinovic-Carugo, K, Puehringer, D. | | Deposit date: | 2018-12-20 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A conformation-specific ON-switch for controlling CAR T cells with an orally available drug.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4U53
 
 | | Crystal structure of Deoxynivalenol bound to the yeast 80S ribosome | | Descriptor: | (3beta,7alpha)-3,7,15-trihydroxy-12,13-epoxytrichothec-9-en-8-one, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
