1QBH
 
 | | SOLUTION STRUCTURE OF A BACULOVIRAL INHIBITOR OF APOPTOSIS (IAP) REPEAT | | Descriptor: | INHIBITOR OF APOPTOSIS PROTEIN (2MIHB/C-IAP-1), ZINC ION | | Authors: | Hinds, M.G, Norton, R.S, Vaux, D.L, Day, C.L. | | Deposit date: | 1999-04-20 | | Release date: | 1999-10-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a baculoviral inhibitor of apoptosis (IAP) repeat.
Nat.Struct.Biol., 6, 1999
|
|
1A7M
 
 | | LEUKAEMIA INHIBITORY FACTOR CHIMERA (MH35-LIF), NMR, 20 STRUCTURES | | Descriptor: | LEUKEMIA INHIBITORY FACTOR | | Authors: | Hinds, M.G, Maurer, T, Zhang, J.-G, Nicola, N.A, Norton, R.S. | | Deposit date: | 1998-03-16 | | Release date: | 1999-04-20 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of leukemia inhibitory factor.
J.Biol.Chem., 273, 1998
|
|
1KD6
 
 | | Solution structure of the eukaryotic pore-forming cytolysin equinatoxin II | | Descriptor: | EQUINATOXIN II | | Authors: | Hinds, M.G, Zhang, W, Anderluh, G, Hansen, P.E, Norton, R.S. | | Deposit date: | 2001-11-12 | | Release date: | 2002-02-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the eukaryotic pore-forming cytolysin equinatoxin II: implications for pore formation.
J.Mol.Biol., 315, 2002
|
|
1O0L
 
 | | THE STRUCTURE OF BCL-W REVEALS A ROLE FOR THE C-TERMINAL RESIDUES IN MODULATING BIOLOGICAL ACTIVITY | | Descriptor: | Apoptosis regulator Bcl-W | | Authors: | Hinds, M.G, Lackmann, M, Skea, G.L, Harrison, P.J, Huang, D.C.S, Day, C.L. | | Deposit date: | 2003-02-22 | | Release date: | 2003-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of Bcl-w reveals a role for the C-terminal residues in modulating biological activity
Embo J., 22, 2003
|
|
2LEL
 
 | | Structure of Cu(I)Cu(II)-CopK from Cupriavidus metallidurans CH34 | | Descriptor: | COPPER (I) ION, COPPER (II) ION, Copper resistance protein K | | Authors: | Hinds, M.G, Xiao, Z, Chong, L.X, Wedd, A.G. | | Deposit date: | 2011-06-16 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of the cooperative binding of Cu(I) and Cu(II) to the CopK protein from Cupriavidus metallidurans CH34.
Biochemistry, 50, 2011
|
|
2M05
 
 | |
6FBX
 
 | |
2KKH
 
 | | Structure of the zinc binding domain of the ATPase HMA4 | | Descriptor: | Putative heavy metal transporter, ZINC ION | | Authors: | Zimmerman, M, Clarke, O, Gulbis, J.M, Keizer, D.W, Jarvis, R.S, Cobbett, C.S, Hinds, M.G, Xiao, Z, Wedd, A.G. | | Deposit date: | 2009-06-20 | | Release date: | 2010-01-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Metal binding affinities of Arabidopsis zinc and copper transporters: selectivities match the relative, but not the absolute, affinities of their amino-terminal domains
Biochemistry, 48, 2009
|
|
2KMF
 
 | | Solution Structure of Psb27 from cyanobacterial photosystem II | | Descriptor: | Photosystem II 11 kDa protein | | Authors: | Mabbitt, P.D, Rautureau, G.J.P, Day, C.L, Wilbanks, S.M, Eaton-Rye, J.J, Hinds, M.G. | | Deposit date: | 2009-07-28 | | Release date: | 2009-09-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Psb27 from cyanobacterial photosystem II
Biochemistry, 48, 2009
|
|
2KUA
 
 | |
2L9M
 
 | | Structure of cIAP1 CARD | | Descriptor: | Baculoviral IAP repeat-containing protein 2 | | Authors: | Day, C.L, Rautureau, G.J.P, Hinds, M.G. | | Deposit date: | 2011-02-21 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | CARD-mediated autoinhibition of cIAP1's E3 ligase activity suppresses cell proliferation and migration.
Mol.Cell, 42, 2011
|
|
2LNJ
 
 | |
4B4S
 
 | |
6H1N
 
 | |
7QTX
 
 | | Kaposi sarcoma associated herpes virus (KSHV) encoded apoptosis inhibitor, KsBcl-2 in complex with Puma BH3 | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Bcl-2, ... | | Authors: | Suraweera, C.D, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2022-01-17 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.11598849 Å) | | Cite: | Structural Insight into KsBcl-2 Mediated Apoptosis Inhibition by Kaposi Sarcoma Associated Herpes Virus.
Viruses, 14, 2022
|
|
7QTW
 
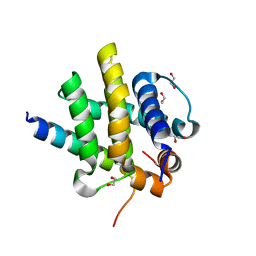 | | Kaposi sarcoma associated herpes virus(KSHV) encoded apoptosis inhibitor, KsBcl-2 in complex with Bid BH3 | | Descriptor: | 1,2-ETHANEDIOL, BH3-interacting domain death agonist p15, Bcl-2 | | Authors: | Suraweera, C.D, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2022-01-17 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural Insight into KsBcl-2 Mediated Apoptosis Inhibition by Kaposi Sarcoma Associated Herpes Virus.
Viruses, 14, 2022
|
|
7ADS
 
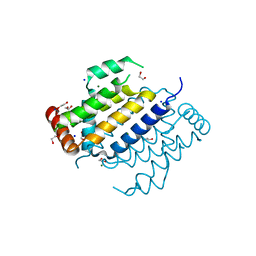 | | Orf virus Apoptosis inhibitor ORFV125 | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis inhibitor, CHLORIDE ION, ... | | Authors: | Suraweera, C.D, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2020-09-16 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.22032046 Å) | | Cite: | Crystal structures of ORFV125 provide insight into orf virus-mediated inhibition of apoptosis.
Biochem.J., 477, 2020
|
|
7ADT
 
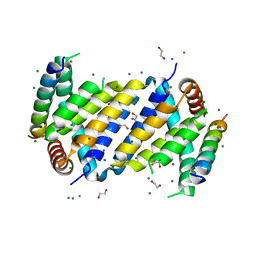 | | Orf virus Apoptosis inhibitor ORFV125 | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis inhibitor, Apoptosis regulator BAX, ... | | Authors: | Suraweera, C.D, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2020-09-16 | | Release date: | 2021-10-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.210037 Å) | | Cite: | Crystal structures of ORFV125 provide insight into orf virus-mediated inhibition of apoptosis.
Biochem.J., 477, 2020
|
|
7P0U
 
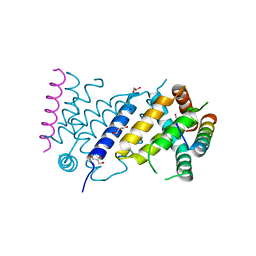 | | ORF virus encoded Bcl-2 homolog ORFV125 in complex with Puma BH3 peptide | | Descriptor: | 1,2-ETHANEDIOL, Activator of apoptosis harakiri, Apoptosis inhibitor, ... | | Authors: | Suraweera, C.D, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2021-06-30 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99374259 Å) | | Cite: | Structural Investigation of Orf Virus Bcl-2 Homolog ORFV125 Interactions with BH3-Motifs from BH3-Only Proteins Puma and Hrk.
Viruses, 13, 2021
|
|
7P0S
 
 | |
7P33
 
 | | Epstein-Barr virus encoded Bcl-2 homolog BHRF-1 in complex with Bid BH3 peptide | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BHRF1, BH3-interacting domain death agonist p15, ... | | Authors: | Suraweera, C.D, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2021-07-07 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.78542733 Å) | | Cite: | Crystal Structures of Epstein-Barr Virus Bcl-2 Homolog BHRF1 Bound to Bid and Puma BH3 Motif Peptides.
Viruses, 14, 2022
|
|
7P9W
 
 | | Epstein-Barr virus encoded apoptosis regulator BHRF1 in complex with Puma BH3 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, AMMONIUM ION, ... | | Authors: | Suraweera, C.D, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.00010061 Å) | | Cite: | Crystal Structures of Epstein-Barr Virus Bcl-2 Homolog BHRF1 Bound to Bid and Puma BH3 Motif Peptides.
Viruses, 14, 2022
|
|
5TWA
 
 | | Crystal structure of Geodia cydonium BHP2 in complex with Lubomirskia baicalensis Bak-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BAK-2 protein, ... | | Authors: | Caria, S, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2016-11-12 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insight into an evolutionarily ancient programmed cell death regulator - the crystal structure of marine sponge BHP2 bound to LB-Bak-2.
Cell Death Dis, 8, 2017
|
|
6WH0
 
 | | Crystal structure of HyBcl-2-4 with HyBax BH3 | | Descriptor: | Apoptosis regulator BAX, Maltodextrin-binding protein,Bcl-2-like 4, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kvansakul, M, Hinds, M.G, Banjara, S. | | Deposit date: | 2020-04-07 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The structural basis of Bcl-2 mediated cell death regulation in hydra.
Biochem.J., 477, 2020
|
|
6WGZ
 
 | |
