9QVM
 
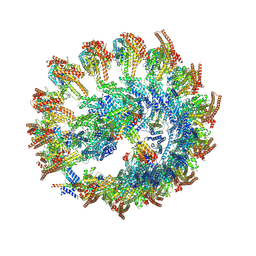 | | Cryo-EM reconstruction of the NEDD1 anchor protein and CDK5RAP2 bound to the gamma-tubulin ring complex | | Descriptor: | Actin, cytoplasmic 1, N-terminally processed, ... | | Authors: | Munoz-Hernandez, H, Xu, Y, Wieczorek, M. | | Deposit date: | 2025-04-11 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Structure of the microtubule-anchoring factor NEDD1 bound to the gamma-tubulin ring complex.
J.Cell Biol., 224, 2025
|
|
9QVN
 
 | | Cryo-EM reconstruction of the NEDD1 anchor protein bound to the gamma-tubulin ring complex | | Descriptor: | Actin, cytoplasmic 1, N-terminally processed, ... | | Authors: | Munoz-Hernandez, H, Xu, Y, Wieczorek, M. | | Deposit date: | 2025-04-11 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structure of the microtubule-anchoring factor NEDD1 bound to the gamma-tubulin ring complex.
J.Cell Biol., 224, 2025
|
|
6QI9
 
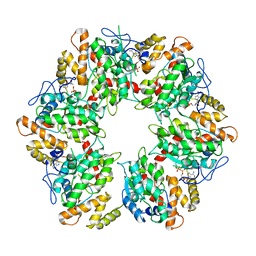 | | Truncated human R2TP complex, structure 4 (ADP-empty) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2 | | Authors: | Munoz-Hernandez, H, Rodriguez, C.F, Llorca, O. | | Deposit date: | 2019-01-18 | | Release date: | 2019-05-15 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.63 Å) | | Cite: | Structural mechanism for regulation of the AAA-ATPases RUVBL1-RUVBL2 in the R2TP co-chaperone revealed by cryo-EM.
Sci Adv, 5, 2019
|
|
6QI8
 
 | | Truncated human R2TP complex, structure 3 (ADP-filled) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2 | | Authors: | Munoz-Hernandez, H, Rodriguez, C.F, Llorca, O. | | Deposit date: | 2019-01-18 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structural mechanism for regulation of the AAA-ATPases RUVBL1-RUVBL2 in the R2TP co-chaperone revealed by cryo-EM.
Sci Adv, 5, 2019
|
|
6FO1
 
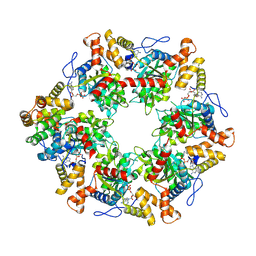 | | Human R2TP subcomplex containing 1 RUVBL1-RUVBL2 hexamer bound to 1 RBD domain from RPAP3. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RNA polymerase II-associated protein 3, RuvB-like 1, ... | | Authors: | Martino, F, Munoz-Hernandez, H, Rodriguez, C.F, Pearl, L.H, Llorca, O. | | Deposit date: | 2018-02-05 | | Release date: | 2018-04-04 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | RPAP3 provides a flexible scaffold for coupling HSP90 to the human R2TP co-chaperone complex.
Nat Commun, 9, 2018
|
|
8CG5
 
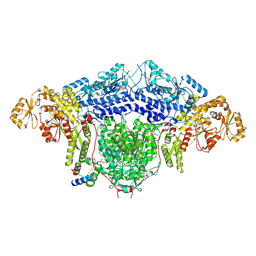 | |
8CG4
 
 | |
8BY8
 
 | |
8CG6
 
 | |
9G40
 
 | | Structure of the Position 7 CMG-decorated gamma-Tubulin Ring Complex from Pig Brain | | Descriptor: | CDK5 regulatory subunit-associated protein 2, Gamma-tubulin complex component, Gamma-tubulin complex component 3, ... | | Authors: | Munoz-Hernandez, H, Krutyholowa, R, Wieczorek, M. | | Deposit date: | 2024-07-12 | | Release date: | 2024-10-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Partial closure of the gamma-tubulin ring complex by CDK5RAP2 activates microtubule nucleation.
Dev.Cell, 59, 2024
|
|
9G3Y
 
 | |
9G3Z
 
 | |
9G3X
 
 | |
1GQ7
 
 | | PROCLAVAMINATE AMIDINO HYDROLASE FROM STREPTOMYCES CLAVULIGERUS | | Descriptor: | MANGANESE (II) ION, PROCLAVAMINATE AMIDINO HYDROLASE | | Authors: | Elkins, J.M, Clifton, I.J, Hernandez, H, Robinson, C.V, Schofield, C.J, Hewitson, K.S. | | Deposit date: | 2001-11-20 | | Release date: | 2002-06-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Oligomeric structure of proclavaminic acid amidino hydrolase: evolution of a hydrolytic enzyme in clavulanic acid biosynthesis.
Biochem. J., 366, 2002
|
|
1GQ6
 
 | | PROCLAVAMINATE AMIDINO HYDROLASE FROM STREPTOMYCES CLAVULIGERUS | | Descriptor: | MANGANESE (II) ION, PROCLAVAMINATE AMIDINO HYDROLASE | | Authors: | Elkins, J.M, Clifton, I.J, Hernandez, H, Robinson, C.V, Schofield, C.J, Hewitson, K.S. | | Deposit date: | 2001-11-20 | | Release date: | 2002-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Oligomeric Structure of Proclavaminic Acid Amidino Hydrolase: Evolution of a Hydrolytic Enzyme in Clavulanic Acid Biosynthesis
Biochem.J., 366, 2002
|
|
2VRD
 
 | | THE STRUCTURE OF THE ZINC FINGER FROM THE HUMAN SPLICEOSOMAL PROTEIN U1C | | Descriptor: | U1 SMALL NUCLEAR RIBONUCLEOPROTEIN C, ZINC ION | | Authors: | Muto, Y, Pomeranz-Krummel, D, Oubridge, C, Hernandez, H, Robinson, C, Neuhaus, D, Nagai, K. | | Deposit date: | 2008-03-31 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure and Biochemical Properties of the Human Spliceosomal Protein U1C
J.Mol.Biol., 341, 2004
|
|
2JAH
 
 | | Biochemical and structural analysis of the Clavulanic acid dehydeogenase (CAD) from Streptomyces clavuligerus | | Descriptor: | CLAVULANIC ACID DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | MacKenzie, A.K, Kershaw, N.J, Hernandez, H, Robinson, C.V, Schofield, C.J, Andersson, I. | | Deposit date: | 2006-11-28 | | Release date: | 2007-02-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Clavulanic Acid Dehydrogenase: Structural and Biochemical Analysis of the Final Step in the Biosynthesis of the Beta-Lactamase Inhibitor Clavulanic Acid
Biochemistry, 46, 2007
|
|
2JAP
 
 | | Clavulanic Acid Dehydrogenase: Structural and Biochemical Analysis of the Final Step in the Biosynthesis of the beta-Lactamase Inhibitor Clavulanic acid | | Descriptor: | (2R,3Z,5R)-3-(2-HYDROXYETHYLIDENE)-7-OXO-4-OXA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, CLAVALDEHYDE DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | MacKenzie, A.K, Kershaw, N.J, Hernandez, H, Robinson, C.V, Schofield, C.J, Andersson, I. | | Deposit date: | 2006-11-29 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Clavulanic Acid Dehydrogenase: Structural and Biochemical Analysis of the Final Step in the Biosynthesis of the Beta-Lactamase Inhibitor Clavulanic Acid
Biochemistry, 46, 2007
|
|
5D6C
 
 | | Structure of 4497 Fab bound to synthetic wall teichoic acid fragment | | Descriptor: | 4-O-[2-acetamido-2-deoxy-beta-D-glucopyranosyl]-5-O-phosphono-D-ribitol, 4497 antibody IgG1 (VH and CH1), 4497 antibody IgK (VL and CL), ... | | Authors: | Lupardus, P.J, Fong, R. | | Deposit date: | 2015-08-12 | | Release date: | 2015-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Novel antibody-antibiotic conjugate eliminates intracellular S. aureus.
Nature, 527, 2015
|
|
7OLE
 
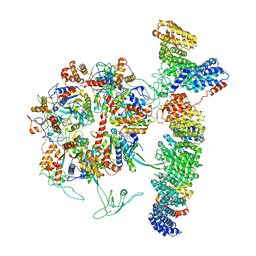 | | Cryo-EM structure of the TELO2-TTI1-TTI2-RUVBL1-RUVBL2 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2, ... | | Authors: | Pal, M, Llorca, O, Pearl, L. | | Deposit date: | 2021-05-19 | | Release date: | 2021-07-07 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structure of the TELO2-TTI1-TTI2 complex and its function in TOR recruitment to the R2TP chaperone.
Cell Rep, 36, 2021
|
|
7ZMC
 
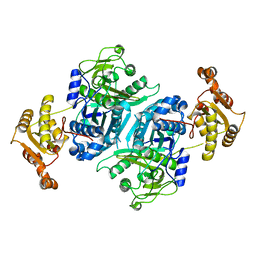 | |
7ZMA
 
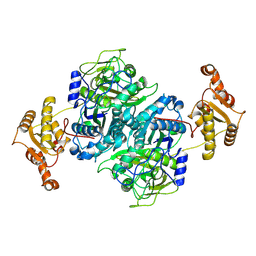 | |
7ZM9
 
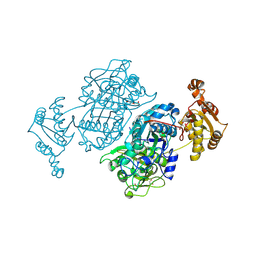 | | Ketosynthase domain 3 of Brevibacillus Brevis orphan BGC11 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Putative polyketide synthase | | Authors: | Tittes, Y.U, Herbst, D.A, Jakob, R.P, Maier, T. | | Deposit date: | 2022-04-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The structure of a polyketide synthase bimodule core.
Sci Adv, 8, 2022
|
|
7ZMF
 
 | | Dehydratase domain of module 3 from Brevibacillus Brevis orphan BGC11 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Putative polyketide synthase | | Authors: | Tittes, Y.U, Herbst, D.A, Jakob, R.P, Maier, T. | | Deposit date: | 2022-04-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The structure of a polyketide synthase bimodule core.
Sci Adv, 8, 2022
|
|
7ZMD
 
 | |
