1AYG
 
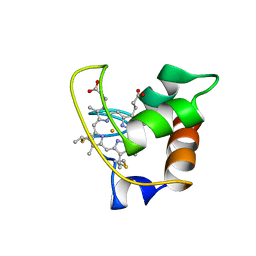 | | SOLUTION STRUCTURE OF CYTOCHROME C-552, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C-552, HEME C | | Authors: | Hasegawa, J, Yoshida, T, Yamazaki, T, Sambongi, Y, Yu, Y, Igarashi, Y, Kodama, T, Yamazaki, K, Hakusui, H, Kyogoku, Y, Kobayashi, Y. | | Deposit date: | 1997-11-04 | | Release date: | 1998-11-25 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of thermostable cytochrome c-552 from Hydrogenobacter thermophilus determined by 1H-NMR spectroscopy.
Biochemistry, 37, 1998
|
|
1DVV
 
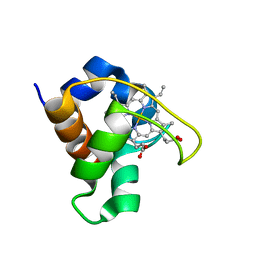 | | SOLUTION STRUCTURE OF THE QUINTUPLE MUTANT OF CYTOCHROME C-551 FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | CYTOCHROME C551, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hasegawa, J, Uchiyama, S, Tanimoto, Y, Mizutani, M, Kobayashi, Y, Sambongi, Y, Igarashi, Y. | | Deposit date: | 2000-01-22 | | Release date: | 2000-11-29 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Selected mutations in a mesophilic cytochrome c confer the stability of a thermophilic counterpart.
J.Biol.Chem., 275, 2000
|
|
2AI5
 
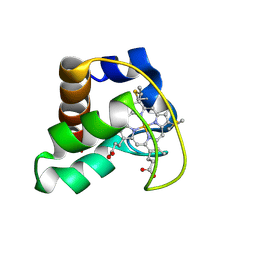 | | Solution Structure of Cytochrome C552, determined by Distributed Computing Implementation for NMR data | | Descriptor: | Cytochrome c-552, HEME C | | Authors: | Nakamura, S, Ichiki, S.I, Takashima, H, Uchiyama, S, Hasegawa, J, Kobayashi, Y, Sambongi, Y, Ohkubo, T. | | Deposit date: | 2005-07-29 | | Release date: | 2006-05-23 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structure of Cytochrome c552 from a Moderate Thermophilic Bacterium, Hydrogenophilus thermoluteolus: Comparative Study on the Thermostability of Cytochrome c
Biochemistry, 45, 2006
|
|
1IW4
 
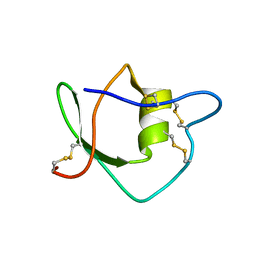 | | Solution structure of ascidian trypsin inhibitor | | Descriptor: | trypsin inhibitor | | Authors: | Hemmi, H, Yoshida, T, Kumazaki, T, Nemoto, N, Hasegawa, J, Nishioka, F, Kyogoku, Y, Yokosawa, H, Kobayashi, Y. | | Deposit date: | 2002-04-19 | | Release date: | 2002-08-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ascidian trypsin inhibitor determined by nuclear magnetic resonance spectroscopy.
Biochemistry, 41, 2002
|
|
2D0S
 
 | | Crystal structure of the Cytochrome C552 from moderate thermophilic bacterium, hydrogenophilus thermoluteolus | | Descriptor: | HEME C, cytochrome c | | Authors: | Nakamura, S, Ichiki, S.I, Takashima, H, Uchiyama, S, Hasegawa, J, Kobayashi, Y, Sambongi, Y, Ohkubo, T. | | Deposit date: | 2005-08-08 | | Release date: | 2006-05-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Cytochrome c552 from a Moderate Thermophilic Bacterium, Hydrogenophilus thermoluteolus: Comparative Study on the Thermostability of Cytochrome c
Biochemistry, 45, 2006
|
|
3B0F
 
 | | Crystal structure of the UBA domain of p62 and its interaction with ubiquitin | | Descriptor: | SULFATE ION, Sequestosome-1 | | Authors: | Isogai, S, Morimoto, D, Arita, K, Unzai, S, Tenno, T, Hasegawa, J, Sou, Y, Komatsu, M, Tanaka, K, Shirakawa, M, Tochio, H. | | Deposit date: | 2011-06-09 | | Release date: | 2011-06-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the ubiquitin-associated (UBA) domain of p62 and its interaction with ubiquitin.
J.Biol.Chem., 286, 2011
|
|
2RRU
 
 | | Solution structure of the UBA omain of p62 and its interaction with ubiquitin | | Descriptor: | Sequestosome-1 | | Authors: | Isogai, S, Morimoto, D, Arita, K, Unzai, S, Tenno, T, Hasegawa, J, Sou, Y, Komatsu, M, Tanaka, K, Shirakawa, M, Tochio, H. | | Deposit date: | 2011-06-09 | | Release date: | 2011-06-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Crystal structure of the UBA omain of p62 and its interaction with ubiquitin
To be Published
|
|
3WDZ
 
 | | Crystal Structure of Keap1 in Complex with phosphorylated p62 | | Descriptor: | Kelch-like ECH-associated protein 1, Peptide from Sequestosome-1 | | Authors: | Fukutomi, T, Takagi, K, Mizushima, T, Tanaka, K, Komatsu, M, Yamamoto, M. | | Deposit date: | 2013-06-26 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy.
Mol.Cell, 51, 2013
|
|
1YNR
 
 | | Crystal structure of the cytochrome c-552 from Hydrogenobacter thermophilus at 2.0 resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cytochrome c-552, HEME C, ... | | Authors: | Travaglini-Allocatelli, C, Gianni, S, Dubey, V.K, Borgia, A, Di Matteo, A, Bonivento, D, Cutruzzola, F, Bren, K.L, Brunori, M. | | Deposit date: | 2005-01-25 | | Release date: | 2005-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An Obligatory Intermediate in the Folding Pathway of Cytochrome c552 from Hydrogenobacter thermophilus
J.Biol.Chem., 280, 2005
|
|
3AJ4
 
 | | Crystal structure of the PH domain of Evectin-2 from human complexed with O-phospho-L-serine | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHOSERINE, Pleckstrin homology domain-containing family B member 2 | | Authors: | Okazaki, S, Kato, R, Wakatsuki, S. | | Deposit date: | 2010-05-21 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Intracellular phosphatidylserine is essential for retrograde membrane traffic through endosomes
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2EXV
 
 | | Crystal structure of the F7A mutant of the cytochrome c551 from Pseudomonas aeruginosa | | Descriptor: | ACETIC ACID, Cytochrome c-551, HEME C | | Authors: | Bonivento, D, Di Matteo, A, Borgia, A, Travaglini-Allocatelli, C, Brunori, M. | | Deposit date: | 2005-11-09 | | Release date: | 2006-02-07 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Unveiling a Hidden Folding Intermediate in c-Type Cytochromes by Protein Engineering
J.Biol.Chem., 281, 2006
|
|
