6LII
 
 | | A quinone oxidoreductase | | Descriptor: | Synaptic vesicle membrane protein VAT-1 homolog | | Authors: | Hakoshima, T, Kim, S.-Y, Mori, T. | | Deposit date: | 2019-12-11 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into vesicle amine transport-1 (VAT-1) as a member of the NADPH-dependent quinone oxidoreductase family.
Sci Rep, 11, 2021
|
|
6LHR
 
 | |
2YVC
 
 | |
1NPM
 
 | | NEUROPSIN, A SERINE PROTEASE EXPRESSED IN THE LIMBIC SYSTEM OF MOUSE BRAIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NEUROPSIN | | Authors: | Kishi, T, Kato, M, Shimizu, T, Kato, K, Matsumoto, K, Yoshida, S, Shiosaka, S, Hakoshima, T. | | Deposit date: | 1998-01-07 | | Release date: | 1999-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of neuropsin, a hippocampal protease involved in kindling epileptogenesis.
J.Biol.Chem., 274, 1999
|
|
2D10
 
 | |
2D11
 
 | |
2A0B
 
 | | HISTIDINE-CONTAINING PHOSPHOTRANSFER DOMAIN OF ARCB FROM ESCHERICHIA COLI | | Descriptor: | HPT DOMAIN, ZINC ION | | Authors: | Kato, M, Mizuno, T, Shimizu, T, Hakoshima, T. | | Deposit date: | 1998-04-02 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Refined structure of the histidine-containing phosphotransfer (HPt) domain of the anaerobic sensor kinase ArcB from Escherichia coli at 1.57 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1A0B
 
 | | HISTIDINE-CONTAINING PHOSPHOTRANSFER DOMAIN OF ARCB FROM ESCHERICHIA COLI | | Descriptor: | AEROBIC RESPIRATION CONTROL SENSOR PROTEIN ARCB, ZINC ION | | Authors: | Kato, M, Mizuno, T, Shimizu, T, Hakoshima, T. | | Deposit date: | 1997-11-27 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Insights into multistep phosphorelay from the crystal structure of the C-terminal HPt domain of ArcB.
Cell(Cambridge,Mass.), 88, 1997
|
|
2IRF
 
 | | CRYSTAL STRUCTURE OF AN IRF-2/DNA COMPLEX. | | Descriptor: | DNA (5'-D(*TP*TP*CP*AP*CP*TP*TP*TP*CP*AP*CP*(5IU)P*T)-3'), DNA (5'-D(P*AP*AP*GP*TP*GP*AP*AP*AP*GP*(5IU)P*GP*A)-3'), INTERFERON REGULATORY FACTOR 2, ... | | Authors: | Fujii, Y, Shimizu, T, Kusumoto, M, Kyogoku, Y, Taniguchi, T, Hakoshima, T. | | Deposit date: | 1999-05-30 | | Release date: | 1999-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of an IRF-DNA complex reveals novel DNA recognition and cooperative binding to a tandem repeat of core sequences.
EMBO J., 18, 1999
|
|
1J19
 
 | | Crystal structure of the radxin FERM domain complexed with the ICAM-2 cytoplasmic peptide | | Descriptor: | 16-mer peptide from Intercellular adhesion molecule-2, radixin | | Authors: | Hamada, K, Shimizu, T, Yonemura, S, Tsukita, S, Tsukita, S, Hakoshima, T. | | Deposit date: | 2002-12-02 | | Release date: | 2003-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of adhesion-molecule recognition by ERM proteins revealed by the crystal structure of the radixin-ICAM-2 complex
EMBO J., 22, 2003
|
|
1GD2
 
 | | CRYSTAL STRUCTURE OF BZIP TRANSCRIPTION FACTOR PAP1 BOUND TO DNA | | Descriptor: | DNA (5'-D(*AP*GP*GP*TP*TP*AP*CP*GP*TP*AP*AP*CP*C)-3'), TRANSCRIPTION FACTOR PAP1 | | Authors: | Fujii, Y, Shimizu, T, Toda, T, Yanagida, M, Hakoshima, T. | | Deposit date: | 2000-08-25 | | Release date: | 2000-10-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the diversity of DNA recognition by bZIP transcription factors.
Nat.Struct.Biol., 7, 2000
|
|
1GC7
 
 | | CRYSTAL STRUCTURE OF THE RADIXIN FERM DOMAIN | | Descriptor: | RADIXIN | | Authors: | Hamada, K, Shimizu, T, Matsui, T, Tsukita, S, Tsukita, S, Hakoshima, T. | | Deposit date: | 2000-07-21 | | Release date: | 2000-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of the membrane-targeting and unmasking mechanisms of the radixin FERM domain.
EMBO J., 19, 2000
|
|
1GC6
 
 | | CRYSTAL STRUCTURE OF THE RADIXIN FERM DOMAIN COMPLEXED WITH INOSITOL-(1,4,5)-TRIPHOSPHATE | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, RADIXIN | | Authors: | Hamada, K, Shimizu, T, Matsui, T, Tsukita, S, Tsukita, S, Hakoshima, T. | | Deposit date: | 2000-07-21 | | Release date: | 2000-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of the membrane-targeting and unmasking mechanisms of the radixin FERM domain.
EMBO J., 19, 2000
|
|
6KYW
 
 | | S8-mSRK-S8-SP11 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor protein kinase SRK8, S locus protein 11 | | Authors: | Murase, K, Hakoshima, T, Mori, T. | | Deposit date: | 2019-09-20 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.60112739 Å) | | Cite: | Mechanism of self/nonself-discrimination in Brassica self-incompatibility.
Nat Commun, 11, 2020
|
|
1CXZ
 
 | | CRYSTAL STRUCTURE OF HUMAN RHOA COMPLEXED WITH THE EFFECTOR DOMAIN OF THE PROTEIN KINASE PKN/PRK1 | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, PROTEIN (HIS-TAGGED TRANSFORMING PROTEIN RHOA(0-181)), ... | | Authors: | Maesaki, R, Ihara, K, Shimizu, T, Kuroda, S, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 1999-08-31 | | Release date: | 1999-10-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structural basis of Rho effector recognition revealed by the crystal structure of human RhoA complexed with the effector domain of PKN/PRK1.
Mol.Cell, 4, 1999
|
|
5XAV
 
 | | Structure of PhaC from Chromobacterium sp. USM2 | | Descriptor: | Intracellular polyhydroxyalkanoate synthase | | Authors: | Chek, M.F, Kim, S.Y, Mori, T, Arsad, H, Samian, M.R, Sudesh, K, Hakoshima, T. | | Deposit date: | 2017-03-15 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.479 Å) | | Cite: | Structure of polyhydroxyalkanoate (PHA) synthase PhaC from Chromobacterium sp. USM2, producing biodegradable plastics
Sci Rep, 7, 2017
|
|
9KNK
 
 | | Crystal structure of full-length PHA synthase (PhaC) from Aeromonas caviae | | Descriptor: | PHA synthase | | Authors: | Chek, M.F, Kim, S.Y, Mori, T, Hakoshima, T. | | Deposit date: | 2024-11-19 | | Release date: | 2025-05-21 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structures of Polyhydroxyalkanoate Synthase PhaC from Aeromonas caviae, Producing Biodegradable Plastics.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9KNL
 
 | | Crystal structure of triethylene glycol-bound full-length PHA synthase (PhaC) from Aeromonas caviae | | Descriptor: | PHA synthase, TRIETHYLENE GLYCOL | | Authors: | Chek, M.F, Kim, S.Y, Mori, T, Hakoshima, T. | | Deposit date: | 2024-11-19 | | Release date: | 2025-05-21 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of Polyhydroxyalkanoate Synthase PhaC from Aeromonas caviae, Producing Biodegradable Plastics.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9KNJ
 
 | | Crystal structure of glycerol-bound full-length PHA synthase (PhaC) from Aeromonas caviae | | Descriptor: | GLYCEROL, PHA synthase | | Authors: | Chek, M.F, Kim, S.Y, Mori, T, Hakoshima, T. | | Deposit date: | 2024-11-19 | | Release date: | 2025-05-21 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of Polyhydroxyalkanoate Synthase PhaC from Aeromonas caviae, Producing Biodegradable Plastics.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
2RRD
 
 | | Structure of HRDC domain from human Bloom syndrome protein, BLM | | Descriptor: | HRDC domain from Bloom syndrome protein | | Authors: | Sato, A, Mishima, M, Nagai, A, Kim, S.Y, Ito, Y, Hakoshima, T, Jee, J.G, Kitano, K. | | Deposit date: | 2010-07-19 | | Release date: | 2010-09-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HRDC domain of human Bloom syndrome protein BLM
J.Biochem., 148, 2010
|
|
6L0W
 
 | | Structure of RLD2 BRX domain bound to LZY3 CCL motif | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, NGR2, ... | | Authors: | Hirano, Y, Futrutani, M, Nishimura, T, Taniguchi, M, Morita, M.T, Hakoshima, T. | | Deposit date: | 2019-09-27 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Polar recruitment of RLD by LAZY1-like protein during gravity signaling in root branch angle control.
Nat Commun, 11, 2020
|
|
4ZKF
 
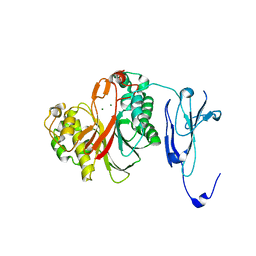 | | Crystal structure of human phosphodiesterase 12 | | Descriptor: | 2',5'-phosphodiesterase 12, MAGNESIUM ION | | Authors: | Kim, S.Y, Kohno, T, Mori, T, Kitano, K, Hakoshima, T. | | Deposit date: | 2015-04-30 | | Release date: | 2015-06-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of human phosphodiesterase
To Be Published
|
|
1QQ2
 
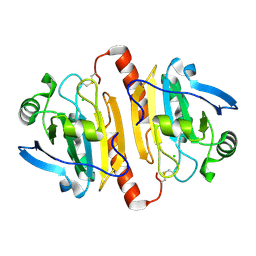 | | CRYSTAL STRUCTURE OF A MAMMALIAN 2-CYS PEROXIREDOXIN, HBP23. | | Descriptor: | CHLORIDE ION, THIOREDOXIN PEROXIDASE 2 | | Authors: | Hirotsu, S, Abe, Y, Okada, K, Nagahara, N, Hori, H, Nishino, T, Hakoshima, T. | | Deposit date: | 1999-06-10 | | Release date: | 1999-10-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a multifunctional 2-Cys peroxiredoxin heme-binding protein 23 kDa/proliferation-associated gene product.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1BDJ
 
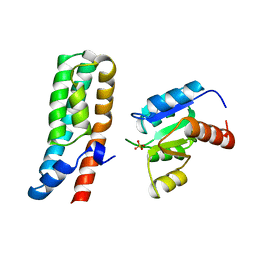 | | COMPLEX STRUCTURE OF HPT DOMAIN AND CHEY | | Descriptor: | AEROBIC RESPIRATION CONTROL SENSOR PROTEIN ARCB, CHEY, SULFATE ION | | Authors: | Kato, M, Mizuno, T, Shimizu, T, Hakoshima, T. | | Deposit date: | 1998-05-10 | | Release date: | 1999-05-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure of the histidine-containing phosphotransfer (HPt) domain of the anaerobic sensor protein ArcB complexed with the chemotaxis response regulator CheY.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
3X23
 
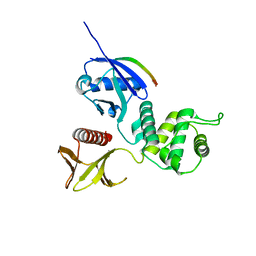 | | Radixin complex | | Descriptor: | Peptide from Matrix metalloproteinase-14, Radixin | | Authors: | Terawaki, S, Kitano, K, Aoyama, M, Mori, T, Hakoshima, T. | | Deposit date: | 2014-12-09 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | MT1-MMP recognition by ERM proteins and its implication in CD44 shedding
Genes Cells, 20, 2015
|
|
