5MJE
 
 | | Crystal structure of the HigB2 toxin in complex with Nb8 | | Descriptor: | Cytotoxic translational repressor of toxin-antitoxin stability system, DI(HYDROXYETHYL)ETHER, Nanobody 8, ... | | Authors: | Hadzi, S, Loris, R. | | Deposit date: | 2016-11-30 | | Release date: | 2017-04-05 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Ribosome-dependent Vibrio cholerae mRNAse HigB2 is regulated by a beta-strand sliding mechanism.
Nucleic Acids Res., 45, 2017
|
|
8A0W
 
 | |
5J9I
 
 | |
5JA9
 
 | | Crystal structure of the HigB2 toxin in complex with Nb6 | | Descriptor: | 1,2-ETHANEDIOL, Nanobody 6, SULFATE ION, ... | | Authors: | Hadzi, S, Loris, R. | | Deposit date: | 2016-04-12 | | Release date: | 2017-04-05 | | Last modified: | 2017-05-17 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Ribosome-dependent Vibrio cholerae mRNAse HigB2 is regulated by a beta-strand sliding mechanism.
Nucleic Acids Res., 45, 2017
|
|
5JAA
 
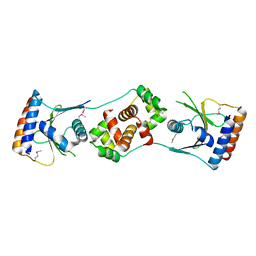 | |
5JA8
 
 | | Crystal structure of the HigB2 toxin in complex with Nb2 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Hadzi, S, Loris, R. | | Deposit date: | 2016-04-12 | | Release date: | 2017-04-05 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Ribosome-dependent Vibrio cholerae mRNAse HigB2 is regulated by a beta-strand sliding mechanism.
Nucleic Acids Res., 45, 2017
|
|
8A0X
 
 | |
7A4Y
 
 | |
7A4T
 
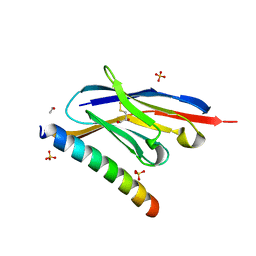 | | Crystal structure of the GCN coiled-coil in complex with nanobody Nb39 | | Descriptor: | 1,2-ETHANEDIOL, ACETYL GROUP, GCN4 isoform 1, ... | | Authors: | Hadzi, S. | | Deposit date: | 2020-08-20 | | Release date: | 2021-05-05 | | Last modified: | 2021-08-11 | | Method: | X-RAY DIFFRACTION (2.124 Å) | | Cite: | A nanobody toolbox targeting dimeric coiled-coil modules for functionalization of designed protein origami structures.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7A50
 
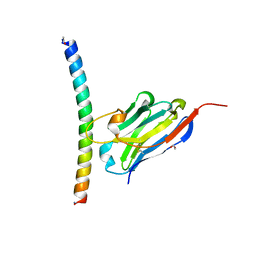 | | Crystal structure of the APH coiled-coil in complex with nanobody Nb26 | | Descriptor: | 1,2-ETHANEDIOL, Coiled-coil APH, Nanobody Nb26 | | Authors: | Hadzi, S. | | Deposit date: | 2020-08-20 | | Release date: | 2021-05-05 | | Last modified: | 2021-08-11 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | A nanobody toolbox targeting dimeric coiled-coil modules for functionalization of designed protein origami structures.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7A48
 
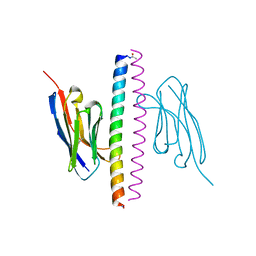 | | Crystal structure of the APH coiled-coil in complex with Nb49 | | Descriptor: | APH colied-coil, Nanobody 49 | | Authors: | Hadzi, S. | | Deposit date: | 2020-08-19 | | Release date: | 2021-05-05 | | Last modified: | 2021-08-11 | | Method: | X-RAY DIFFRACTION (1.547 Å) | | Cite: | A nanobody toolbox targeting dimeric coiled-coil modules for functionalization of designed protein origami structures.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7A4D
 
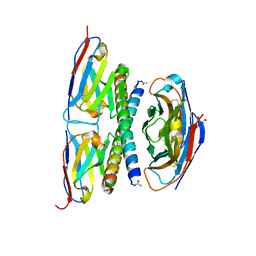 | |
7OWX
 
 | |
8P4Y
 
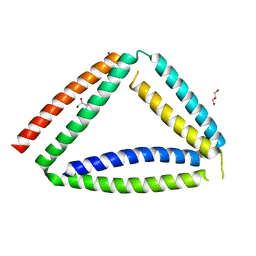 | | Coiled-coil protein origami triangle | | Descriptor: | GLYCEROL, Protein origami triangle | | Authors: | Satler, T, Hadzi, S, Jerala, R. | | Deposit date: | 2023-05-23 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Crystal Structure of de Novo Designed Coiled-Coil Protein Origami Triangle.
J.Am.Chem.Soc., 145, 2023
|
|
8BT1
 
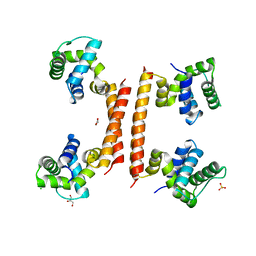 | | YdaT transcription regulator (CII functional analog) | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Prolic-Kalinsek, M, Loris, R. | | Deposit date: | 2022-11-27 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (2.39788437 Å) | | Cite: | Structural basis of DNA binding by YdaT, a functional equivalent of the CII repressor in the cryptic prophage CP-933P from Escherichia coli O157:H7.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6F8S
 
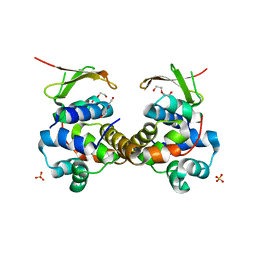 | | Toxin-Antitoxin complex GraTA | | Descriptor: | DI(HYDROXYETHYL)ETHER, Putative Killer protein, SULFATE ION, ... | | Authors: | Talavera, A, Loris, R. | | Deposit date: | 2017-12-13 | | Release date: | 2018-07-11 | | Last modified: | 2019-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A dual role in regulation and toxicity for the disordered N-terminus of the toxin GraT.
Nat Commun, 10, 2019
|
|
6F8H
 
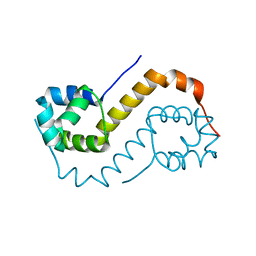 | | antitoxin GraA | | Descriptor: | XRE family transcriptional regulator | | Authors: | Talavera, A, Loris, R. | | Deposit date: | 2017-12-13 | | Release date: | 2019-01-30 | | Last modified: | 2019-03-13 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | A dual role in regulation and toxicity for the disordered N-terminus of the toxin GraT.
Nat Commun, 10, 2019
|
|
6FIX
 
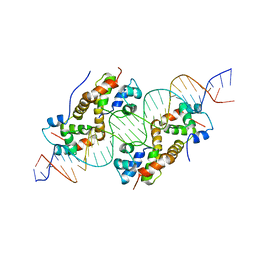 | | antitoxin GraA in complex with its operator | | Descriptor: | DNA (30-MER), XRE family transcriptional regulator | | Authors: | Talavera, A, Loris, R. | | Deposit date: | 2018-01-19 | | Release date: | 2019-01-30 | | Last modified: | 2019-03-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | A dual role in regulation and toxicity for the disordered N-terminus of the toxin GraT.
Nat Commun, 10, 2019
|
|
8C7K
 
 | |
8CO2
 
 | |
