2OPA
 
 | | YwhB binary complex with 2-Fluoro-p-hydroxycinnamate | | Descriptor: | 2-FLUORO-3-(4-HYDROXYPHENYL)-2E-PROPENEOATE, Probable tautomerase ywhB | | Authors: | Hackert, M.L, Whitman, C.P, Almrud, J.J. | | Deposit date: | 2007-01-28 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of YwhB, a 4-Oxalocrotonate Tautomerase Homologue from Bacillus subtilis: the Structural Basis for Catalysis, Inhibition, and Reaction Stereoselectivity.
TO BE PUBLISHED
|
|
2OP8
 
 | |
1ORD
 
 | | CRYSTALLOGRAPHIC STRUCTURE OF A PLP-DEPENDENT ORNITHINE DECARBOXYLASE FROM LACTOBACILLUS 30A TO 3.1 ANGSTROMS RESOLUTION | | Descriptor: | ORNITHINE DECARBOXYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Hackert, M.L, Momany, C, Ernst, S, Ghosh, R. | | Deposit date: | 1995-02-08 | | Release date: | 1995-09-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic structure of a PLP-dependent ornithine decarboxylase from Lactobacillus 30a to 3.0 A resolution.
J.Mol.Biol., 252, 1995
|
|
1HLB
 
 | |
2ORM
 
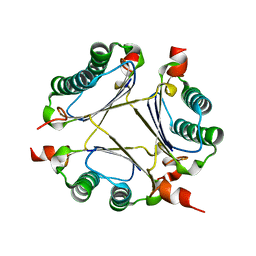 | | Crystal Structure of the 4-Oxalocrotonate Tautomerase Homologue DmpI from Helicobacter pylori. | | Descriptor: | Probable tautomerase HP0924 | | Authors: | Hackert, M.L, Whitman, C.P, Almrud, J.J, Dasgupta, R, Czerwinski, R.M, Kern, A.D. | | Deposit date: | 2007-02-03 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and structural characterization of DmpI from Helicobacter pylori and Archaeoglobus fulgidus, two 4-oxalocrotonate tautomerase family members.
Bioorg.Chem., 38, 2010
|
|
3M20
 
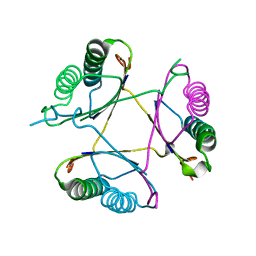 | | Crystal structure of DmpI from Archaeoglobus fulgidus determined to 2.37 Angstroms resolution | | Descriptor: | 4-oxalocrotonate tautomerase, putative | | Authors: | Hackert, M.L, Whitman, C.P, Almrud, J.J, Dasgupta, R, Kern, A.D. | | Deposit date: | 2010-03-06 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Kinetic and structural characterization of DmpI from Helicobacter pylori and Archaeoglobus fulgidus, two 4-oxalocrotonate tautomerase family members.
Bioorg.Chem., 38, 2010
|
|
3M21
 
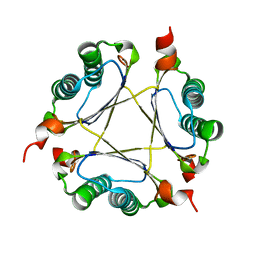 | | Crystal structure of DmpI from Helicobacter pylori Determined to 1.9 Angstroms resolution | | Descriptor: | Probable tautomerase HP_0924 | | Authors: | Hackert, M.L, Whitman, C.P, Almrud, J.J, Dasgupta, R, Kern, A.D, Czerwinski, R.M. | | Deposit date: | 2010-03-06 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and structural characterization of DmpI from Helicobacter pylori and Archaeoglobus fulgidus, two 4-oxalocrotonate tautomerase family members.
Bioorg.Chem., 38, 2010
|
|
1HLM
 
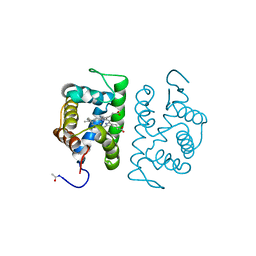 | |
1C4T
 
 | | CATALYTIC DOMAIN FROM TRIMERIC DIHYDROLIPOAMIDE SUCCINYLTRANSFERASE | | Descriptor: | PROTEIN (DIHYDROLIPOAMIDE SUCCINYLTRANSFERASE), SULFATE ION | | Authors: | Knapp, J.E, Carroll, D, Lawson, J.E, Ernst, S.R, Reed, L.J, Hackert, M.L. | | Deposit date: | 1999-09-22 | | Release date: | 2000-02-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Expression, purification, and structural analysis of the trimeric form of the catalytic domain of the Escherichia coli dihydrolipoamide succinyltransferase.
Protein Sci., 9, 2000
|
|
1PYA
 
 | |
7ODC
 
 | | CRYSTAL STRUCTURE ORNITHINE DECARBOXYLASE FROM MOUSE, TRUNCATED 37 RESIDUES FROM THE C-TERMINUS, TO 1.6 ANGSTROM RESOLUTION | | Descriptor: | PROTEIN (ORNITHINE DECARBOXYLASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Kern, A.D, Oliveira, M.A, Coffino, P, Hackert, M.L. | | Deposit date: | 1999-03-03 | | Release date: | 1999-10-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of mammalian ornithine decarboxylase at 1.6 A resolution: stereochemical implications of PLP-dependent amino acid decarboxylases.
Structure Fold.Des., 7, 1999
|
|
6V0T
 
 | | Crystal Structure of Catalytic Subunit of Bovine Pyruvate Dehydrogenase Phosphatase 1 - Catalytic Domain | | Descriptor: | MANGANESE (II) ION, SULFATE ION, [Pyruvate dehydrogenase [acetyl-transferring]]-phosphatase 1, ... | | Authors: | Guo, Y, Qiu, W, Ernst, S.R, Carroll, D.W, Hackert, M.L. | | Deposit date: | 2019-11-19 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the catalytic subunit of bovine pyruvate dehydrogenase phosphatase.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
1C4K
 
 | | ORNITHINE DECARBOXYLASE MUTANT (GLY121TYR) | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, PROTEIN (ORNITHINE DECARBOXYLASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Vitali, J, Hackert, M.L. | | Deposit date: | 1999-08-26 | | Release date: | 2000-02-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional structure of the Gly121Tyr dimeric form of ornithine decarboxylase from Lactobacillus 30a.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
3LDH
 
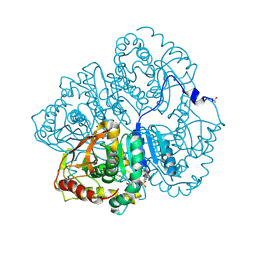 | | A comparison of the structures of apo dogfish m4 lactate dehydrogenase and its ternary complexes | | Descriptor: | LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PYRUVIC ACID | | Authors: | White, J.L, Hackert, M.L, Buehner, M, Adams, M.J, Ford, G.C, Lentzjunior, P.J, Smiley, I.E, Steindel, S.J, Rossmann, M.G. | | Deposit date: | 1974-06-06 | | Release date: | 1977-04-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A comparison of the structures of apo dogfish M4 lactate dehydrogenase and its ternary complexes.
J.Mol.Biol., 102, 1976
|
|
2FM7
 
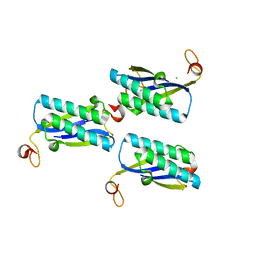 | |
2GDG
 
 | |
1ZO0
 
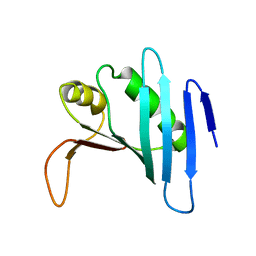 | |
2AAG
 
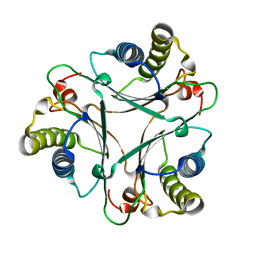 | | Crystal Structures of the Wild-type, Mutant-P1A and Inactivated Malonate Semialdehyde Decarboxylase: A Structural Basis for the Decarboxylase and Hydratase Activities | | Descriptor: | Malonate Semialdehyde Decarboxylase | | Authors: | Almrud, J.J, Poelarends, G.J, Johnson Jr, W.H, Serrano, H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2005-07-13 | | Release date: | 2005-11-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of the Wild-Type, P1A Mutant, and Inactivated Malonate Semialdehyde Decarboxylase: A Structural Basis for the Decarboxylase and Hydratase Activities
Biochemistry, 44, 2005
|
|
2AAL
 
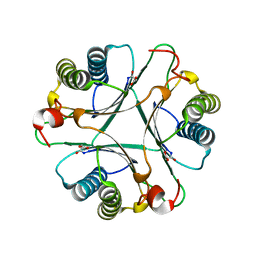 | | Crystal Structures of the Wild-type, Mutant-P1A and Inactivated Malonate Semialdehyde Decarboxylase: A Structural Basis for the Decarboxylase and Hydratase Activities | | Descriptor: | MALONATE ION, Malonate semialdehyde decarboxylase | | Authors: | Almrud, J.J, Poelarends, G.J, Johnson Jr, W.H, Serrano, H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2005-07-13 | | Release date: | 2005-11-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structures of the Wild-Type, P1A Mutant, and Inactivated Malonate Semialdehyde Decarboxylase: A Structural Basis for the Decarboxylase and Hydratase Activities
Biochemistry, 44, 2005
|
|
2AAJ
 
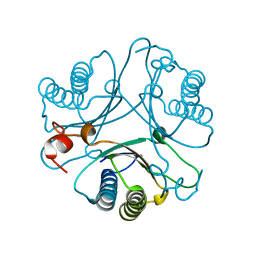 | | Crystal Structures of the Wild-type, Mutant-P1A and Inactivated Malonate Semialdehyde Decarboxylase: A Structural Basis for the Decarboxylase and Hydratase Activities | | Descriptor: | Malonate Semialdehyde Decarboxylase | | Authors: | Almrud, J.J, Poelarends, G.J, Johnson Jr, W.H, Serrano, H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2005-07-13 | | Release date: | 2005-11-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Crystal Structures of the Wild-Type, P1A Mutant, and Inactivated Malonate Semialdehyde Decarboxylase: A Structural Basis for the Decarboxylase and Hydratase Activities
Biochemistry, 44, 2005
|
|
1BJP
 
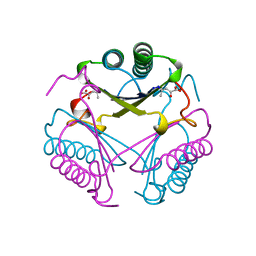 | | CRYSTAL STRUCTURE OF 4-OXALOCROTONATE TAUTOMERASE INACTIVATED BY 2-OXO-3-PENTYNOATE AT 2.4 ANGSTROMS RESOLUTION | | Descriptor: | 2-OXO-3-PENTENOIC ACID, 4-OXALOCROTONATE TAUTOMERASE | | Authors: | Taylor, A.B, Czerwinski, R.M, Johnson Junior, W.H, Whitman, C.P, Hackert, M.L. | | Deposit date: | 1998-06-26 | | Release date: | 1998-12-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of 4-oxalocrotonate tautomerase inactivated by 2-oxo-3-pentynoate at 2.4 A resolution: analysis and implications for the mechanism of inactivation and catalysis.
Biochemistry, 37, 1998
|
|
1D7K
 
 | | CRYSTAL STRUCTURE OF HUMAN ORNITHINE DECARBOXYLASE AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | HUMAN ORNITHINE DECARBOXYLASE | | Authors: | Almrud, J.J, Oliveira, M.A, Kern, A.D, Grishin, N.V, Phillips, M.A, Hackert, M.L. | | Deposit date: | 1999-10-18 | | Release date: | 2000-10-25 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human ornithine decarboxylase at 2.1 A resolution: structural insights to antizyme binding.
J.Mol.Biol., 295, 2000
|
|
4LHP
 
 | | Crystal Structure of Native FG41Malonate Semialdehyde Decarboxylase | | Descriptor: | FG41 Malonate Semialdehyde Decarboxylase, PHOSPHATE ION, SULFATE ION | | Authors: | Guo, Y, Serrano, H, Poelarends, G.J, Johnson Jr, W.H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2013-07-01 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Kinetic, Mutational, and Structural Analysis of Malonate Semialdehyde Decarboxylase from Coryneform Bacterium Strain FG41: Mechanistic Implications for the Decarboxylase and Hydratase Activities.
Biochemistry, 52, 2013
|
|
4LHO
 
 | | Crystal Structure of FG41Malonate Semialdehyde Decarboxylase inhibited by 3-bromopropiolate | | Descriptor: | 3-chloro-3-oxopropanoic acid, FG41 Malonate Semialdehyde Decarboxylase, PHOSPHATE ION | | Authors: | Guo, Y, Serrano, H, Poelarends, G.J, Johnson Jr, W.H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2013-07-01 | | Release date: | 2013-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.224 Å) | | Cite: | Kinetic, Mutational, and Structural Analysis of Malonate Semialdehyde Decarboxylase from Coryneform Bacterium Strain FG41: Mechanistic Implications for the Decarboxylase and Hydratase Activities.
Biochemistry, 52, 2013
|
|
1E2O
 
 | | CATALYTIC DOMAIN FROM DIHYDROLIPOAMIDE SUCCINYLTRANSFERASE | | Descriptor: | DIHYDROLIPOAMIDE SUCCINYLTRANSFERASE, SULFATE ION | | Authors: | Knapp, J.E, Mitchell, D.T, Yazdi, M.A, Ernst, S.R, Reed, L.J, Hackert, M.L. | | Deposit date: | 1998-05-26 | | Release date: | 1998-12-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the truncated cubic core component of the Escherichia coli 2-oxoglutarate dehydrogenase multienzyme complex.
J.Mol.Biol., 280, 1998
|
|
