7RRO
 
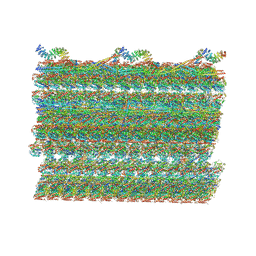 | | Structure of the 48-nm repeat doublet microtubule from bovine tracheal cilia | | Descriptor: | Armadillo repeat containing 4, Chromosome 3 C1orf194 homolog, Cilia and flagella associated protein 161, ... | | Authors: | Gui, M, Anderson, J.R, Botsch, J.J, Meleppattu, S, Singh, S.K, Zhang, Q, Brown, A. | | Deposit date: | 2021-08-10 | | Release date: | 2021-10-27 | | Last modified: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | De novo identification of mammalian ciliary motility proteins using cryo-EM.
Cell, 184, 2021
|
|
5WRG
 
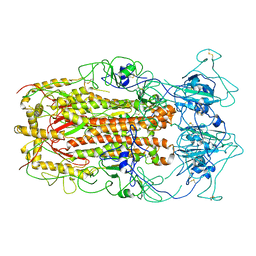 | | SARS-CoV spike glycoprotein | | Descriptor: | Spike glycoprotein | | Authors: | Gui, M, Song, W, Xiang, Y, Wang, X. | | Deposit date: | 2016-12-01 | | Release date: | 2017-01-11 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding.
Cell Res., 27, 2017
|
|
5XLR
 
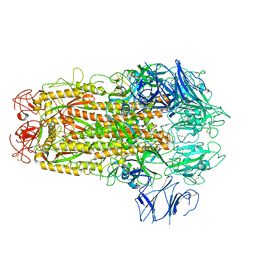 | | Structure of SARS-CoV spike glycoprotein | | Descriptor: | Spike glycoprotein | | Authors: | Gui, M, Song, W, Xiang, Y, Wang, X. | | Deposit date: | 2017-05-11 | | Release date: | 2017-06-07 | | Last modified: | 2019-10-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding.
Cell Res., 27, 2017
|
|
7SQC
 
 | | Ciliary C1 central pair apparatus isolated from Chlamydomonas reinhardtii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CPC1, Calmodulin, ... | | Authors: | Gui, M, Wang, X, Dutcher, S.K, Brown, A, Zhang, R. | | Deposit date: | 2021-11-05 | | Release date: | 2022-04-13 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Ciliary central apparatus structure reveals mechanisms of microtubule patterning.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SOM
 
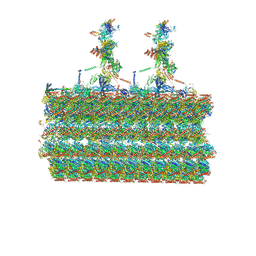 | | Ciliary C2 central pair apparatus isolated from Chlamydomonas reinhardtii | | Descriptor: | Cilia- and flagella-associated protein 20, FAP147, FAP178, ... | | Authors: | Gui, M, Wang, X, Dutcher, S.K, Brown, A, Zhang, R. | | Deposit date: | 2021-11-01 | | Release date: | 2022-04-13 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ciliary central apparatus structure reveals mechanisms of microtubule patterning.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UNG
 
 | | 48-nm repeat of the human respiratory doublet microtubule | | Descriptor: | Cilia- and flagella-associated protein 161, Cilia- and flagella-associated protein 20, Cilia- and flagella-associated protein 45, ... | | Authors: | Gui, M, Croft, J.T, Zabeo, D, Acharya, V, Kollman, J.M, Burgoyne, T, Hoog, J.L, Brown, A. | | Deposit date: | 2022-04-11 | | Release date: | 2022-10-05 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | SPACA9 is a lumenal protein of human ciliary singlet and doublet microtubules.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UN1
 
 | | 8-nm repeat of the human sperm tip singlet microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Gui, M, Croft, J.T, Zabeo, D, Acharya, V, Kollman, J.M, Burgoyne, T, Hoog, J.L, Brown, A. | | Deposit date: | 2022-04-08 | | Release date: | 2022-10-05 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | SPACA9 is a lumenal protein of human ciliary singlet and doublet microtubules.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7JU4
 
 | | Radial spoke 2 stalk, IDAc, and N-DRC attached with doublet microtubule | | Descriptor: | 28 kDa inner dynein arm light chain, axonemal, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gui, M, Ma, M, Sze-Tu, E, Wang, X, Koh, F, Zhong, E, Berger, B, Davis, J, Dutcher, S, Zhang, R, Brown, A. | | Deposit date: | 2020-08-19 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of radial spokes and associated complexes important for ciliary motility.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7JTK
 
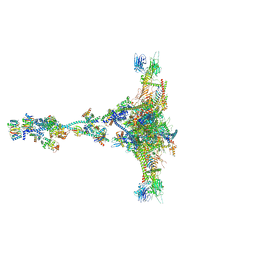 | | Radial spoke 1 isolated from Chlamydomonas reinhardtii | | Descriptor: | Cytochrome b5 heme-binding domain-containing protein, Dynein 8 kDa light chain, flagellar outer arm, ... | | Authors: | Gui, M, Ma, M, Sze-Tu, E, Wang, X, Koh, F, Zhong, E, Berger, B, Davis, J, Dutcher, S, Zhang, R, Brown, A. | | Deposit date: | 2020-08-17 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of radial spokes and associated complexes important for ciliary motility.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7JTS
 
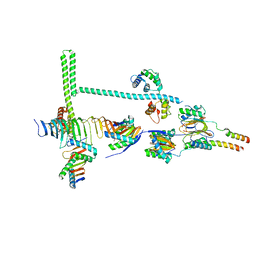 | | Stalk of radial spoke 1 attached with doublet microtubule from Chlamydomonas reinhardtii | | Descriptor: | Calmodulin, Dynein 8 kDa light chain, flagellar outer arm, ... | | Authors: | Gui, M, Ma, M, Sze-Tu, E, Wang, X, Koh, F, Zhong, E, Berger, B, Davis, J, Dutcher, S, Zhang, R, Brown, A. | | Deposit date: | 2020-08-18 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structures of radial spokes and associated complexes important for ciliary motility.
Nat.Struct.Mol.Biol., 28, 2021
|
|
8J07
 
 | |
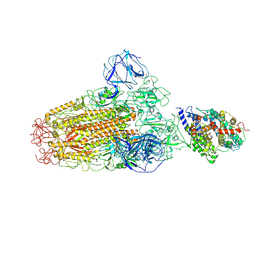
6ACG
 
 | |
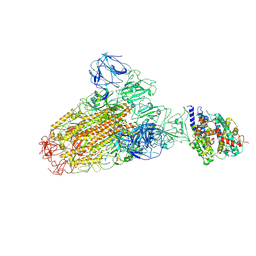
6ACJ
 
 | |
6ACK
 
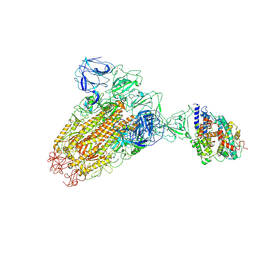 | |
6ACC
 
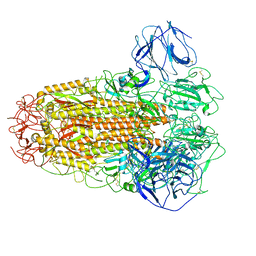 | |
6ACD
 
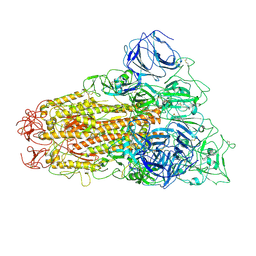 | |
7K3A
 
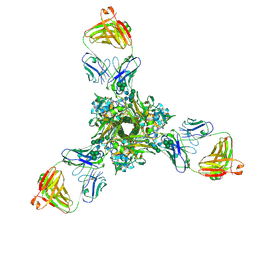 | | Structure of full-length influenza HA with a head-binding antibody at pH 5.2, conformation B, fusion peptide release | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Gui, M, Gao, J, Xiang, Y. | | Deposit date: | 2020-09-10 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural intermediates in the low pH-induced transition of influenza hemagglutinin.
Plos Pathog., 16, 2020
|
|
7K3B
 
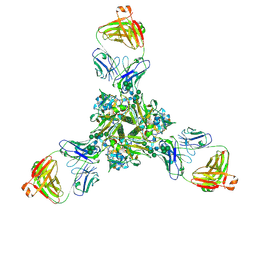 | | Structure of full-length influenza HA with a head-binding antibody at pH 5.2, conformation C, central helices splay | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Gui, M, Gao, J, Xiang, Y. | | Deposit date: | 2020-09-10 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural intermediates in the low pH-induced transition of influenza hemagglutinin.
Plos Pathog., 16, 2020
|
|
7K37
 
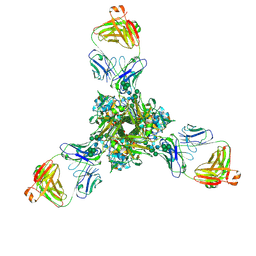 | | Structure of full-length influenza HA with a head-binding antibody at pH 7.8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Gui, M, Xiang, Y, Gao, J. | | Deposit date: | 2020-09-10 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural intermediates in the low pH-induced transition of influenza hemagglutinin.
Plos Pathog., 16, 2020
|
|
7K39
 
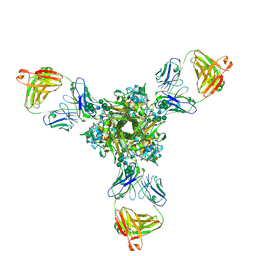 | | Structure of full-length influenza HA with a head-binding antibody at pH 5.2, conformation A, neutral pH-like | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Gui, M, Gao, J, Xiang, Y. | | Deposit date: | 2020-09-10 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural intermediates in the low pH-induced transition of influenza hemagglutinin.
Plos Pathog., 16, 2020
|
|
8IYJ
 
 | | Cryo-EM structure of the 48-nm repeat doublet microtubule from mouse sperm | | Descriptor: | Cilia and flagella-associated protein 77, Cilia- and flagella-associated protein 107, Cilia- and flagella-associated protein 141, ... | | Authors: | Zhou, L.N, Gui, M, Wu, J.P. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of sperm flagellar doublet microtubules expand the genetic spectrum of male infertility.
Cell, 186, 2023
|
|
6VBU
 
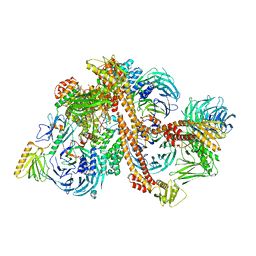 | | Structure of the bovine BBSome complex | | Descriptor: | BBS1 domain-containing protein, Bardet-Biedl syndrome 18 protein, Bardet-Biedl syndrome 2 protein homolog, ... | | Authors: | Singh, S.K, Gui, M, Koh, F, Yip, M.C.J, Brown, A. | | Deposit date: | 2019-12-19 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and activation mechanism of the BBSome membrane protein trafficking complex.
Elife, 9, 2020
|
|
6VBV
 
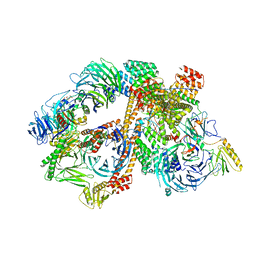 | | Structure of the bovine BBSome:ARL6:GTP complex | | Descriptor: | ADP-ribosylation factor-like protein 6, BBS1 domain-containing protein, Bardet-Biedl syndrome 18 protein, ... | | Authors: | Singh, S.K, Gui, M, Koh, F, Yip, M.C.J, Brown, A. | | Deposit date: | 2019-12-19 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and activation mechanism of the BBSome membrane protein trafficking complex.
Elife, 9, 2020
|
|
5FB5
 
 | |
5FEI
 
 | | Crystal structure of the bacteriophage phi29 tail knob protein gp9 truncation variant | | Descriptor: | DI(HYDROXYETHYL)ETHER, Distal tube protein | | Authors: | Xu, J.W, Gui, M, Wang, D.H, Xiang, Y. | | Deposit date: | 2015-12-17 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | The bacteriophage 29 tail possesses a pore-forming loop for cell membrane penetration.
Nature, 534, 2016
|
|
