1HST
 
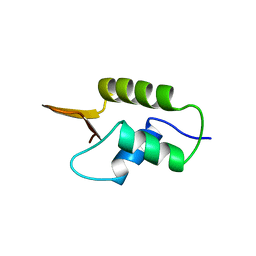 | | CRYSTAL STRUCTURE OF GLOBULAR DOMAIN OF HISTONE H5 AND ITS IMPLICATIONS FOR NUCLEOSOME BINDING | | Descriptor: | HISTONE H5 | | Authors: | Ramakrishnan, V, Finch, J.T, Graziano, V, Lee, P.L, Sweet, R.M. | | Deposit date: | 1993-03-30 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of globular domain of histone H5 and its implications for nucleosome binding.
Nature, 362, 1993
|
|
1P69
 
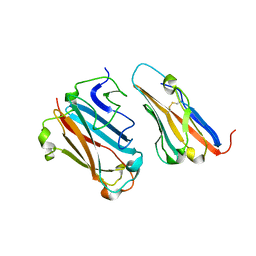 | | STRUCTURAL BASIS FOR VARIATION IN ADENOVIRUS AFFINITY FOR THE CELLULAR RECEPTOR CAR (P417S MUTANT) | | Descriptor: | Coxsackievirus and adenovirus receptor, Fiber protein | | Authors: | Howitt, J, Bewley, M.C, Graziano, V, Flanagan, J.M, Freimuth, P. | | Deposit date: | 2003-04-29 | | Release date: | 2004-05-11 | | Last modified: | 2018-08-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for variation in adenovirus affinity for the cellular coxsackievirus and adenovirus receptor.
J.Biol.Chem., 278, 2003
|
|
1P6A
 
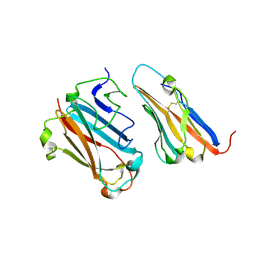 | | STRUCTURAL BASIS FOR VARIATION IN ADENOVIRUS AFFINITY FOR THE CELLULAR RECEPTOR CAR (S489Y MUTANT) | | Descriptor: | Coxsackievirus and adenovirus receptor, Fiber protein | | Authors: | Howitt, J, Bewley, M.C, Graziano, V, Flanagan, J.M, Freimuth, P. | | Deposit date: | 2003-04-29 | | Release date: | 2004-05-11 | | Last modified: | 2018-08-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for variation in adenovirus affinity for the cellular coxsackievirus and adenovirus receptor.
J.Biol.Chem., 278, 2003
|
|
1CI0
 
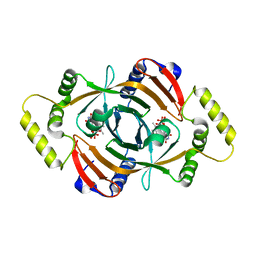 | | PNP OXIDASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | FLAVIN MONONUCLEOTIDE, PROTEIN (PNP OXIDASE) | | Authors: | Shi, W, Ostrov, D.A, Gerchman, S.E, Graziano, V, Kycia, H, Studier, B, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 1999-04-06 | | Release date: | 1999-08-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structure of PNP Oxidase from S. Cerevisiae
To be Published
|
|
1YG6
 
 | | ClpP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit | | Authors: | Bewley, M.C, Graziano, V, Griffin, K, Flanagan, J.M. | | Deposit date: | 2005-01-04 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The asymmetry in the mature amino-terminus of ClpP facilitates a local symmetry match in ClpAP and ClpXP complexes.
J.Struct.Biol., 153, 2006
|
|
1YG8
 
 | |
2B5G
 
 | | Wild Type SSAT- 1.7A structure | | Descriptor: | Diamine acetyltransferase 1, SULFATE ION | | Authors: | Bewley, M.C, Graziano, V, Jiang, J.S, Matz, E, Studier, F.W, Pegg, A.P, Coleman, C.S, Flanagan, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-09-28 | | Release date: | 2006-01-17 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of wild-type and mutant human spermidine/spermine N1-acetyltransferase, a potential therapeutic drug target
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2B58
 
 | | SSAT with coa_sp, spermine disordered, K26R mutant | | Descriptor: | COENZYME A, Diamine acetyltransferase 1 | | Authors: | Bewley, M.C, Graziano, V, Jiang, J.S, Matz, E, Studier, F.W, Pegg, A.P, Coleman, C.S, Flanagan, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-09-27 | | Release date: | 2006-01-24 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of wild-type and mutant human spermidine/spermine N1-acetyltransferase, a potential therapeutic drug target
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2B3V
 
 | | Spermine spermidine acetyltransferase in complex with acetylcoa, K26R mutant | | Descriptor: | ACETYL COENZYME *A, Diamine acetyltransferase 1 | | Authors: | Bewley, M.C, Graziano, V, Jiang, J.S, Matz, E, Studier, F.W, Pegg, A.P, Coleman, C.S, Flanagan, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-09-21 | | Release date: | 2006-01-17 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of wild-type and mutant human spermidine/spermine N1-acetyltransferase, a potential therapeutic drug target.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2B4D
 
 | | SSAT+COA+SP- SP disordered | | Descriptor: | COENZYME A, Diamine acetyltransferase 1 | | Authors: | Bewley, M.C, Graziano, V, Jiang, J.S, Matz, E, Studier, F.W, Pegg, A.P, Coleman, C.S, Flanagan, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-09-23 | | Release date: | 2006-01-17 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of wild-type and mutant human spermidine/spermine N1-acetyltransferase, a potential therapeutic drug target
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2B4B
 
 | | SSAT+COA+BE-3-3-3, K26R mutant | | Descriptor: | COENZYME A, Diamine acetyltransferase 1, N-ETHYL-N-[3-(PROPYLAMINO)PROPYL]PROPANE-1,3-DIAMINE | | Authors: | Bewley, M.C, Graziano, V, Jiang, J.S, Matz, E, Studier, F.W, Pegg, A.P, Coleman, C.S, Flanagan, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-09-23 | | Release date: | 2006-01-17 | | Last modified: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of wild-type and mutant human spermidine/spermine N1-acetyltransferase, a potential therapeutic drug target
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2B3U
 
 | | Human Spermine spermidine acetyltransferase K26R mutant | | Descriptor: | Diamine acetyltransferase 1, SULFATE ION | | Authors: | Bewley, M.C, Graziano, V, Jiang, J.S, Matz, E, Studier, F.W, Pegg, A.P, Coleman, C.S, Flanagan, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-09-21 | | Release date: | 2006-01-17 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of wild-type and mutant human spermidine/spermine N1-acetyltransferase, a potential therapeutic drug target.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
4EKF
 
 | |
1B54
 
 | | CRYSTAL STRUCTURE OF A YEAST HYPOTHETICAL PROTEIN-A STRUCTURE FROM BNL'S HUMAN PROTEOME PROJECT | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, YEAST HYPOTHETICAL PROTEIN | | Authors: | Swaminathan, S, Eswaramoorthy, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 1999-01-12 | | Release date: | 1999-01-27 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a yeast hypothetical protein selected by a structural genomics approach.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1TIG
 
 | | TRANSLATION INITIATION FACTOR 3 C-TERMINAL DOMAIN | | Descriptor: | TRANSLATION INITIATION FACTOR 3 | | Authors: | Biou, V, Shu, F, Ramakrishnan, V. | | Deposit date: | 1995-08-16 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystallography shows that translational initiation factor IF3 consists of two compact alpha/beta domains linked by an alpha-helix.
EMBO J., 14, 1995
|
|
1TIF
 
 | | TRANSLATION INITIATION FACTOR 3 N-TERMINAL DOMAIN | | Descriptor: | CHLORIDE ION, TRANSLATION INITIATION FACTOR 3, TRIMETHYL LEAD ION | | Authors: | Biou, V, Shu, F, Ramakrishnan, V. | | Deposit date: | 1995-08-16 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallography shows that translational initiation factor IF3 consists of two compact alpha/beta domains linked by an alpha-helix.
EMBO J., 14, 1995
|
|
1CT5
 
 | | CRYSTAL STRUCTURE OF YEAST HYPOTHETICAL PROTEIN YBL036C-SELENOMET CRYSTAL | | Descriptor: | PROTEIN (YEAST HYPOTHETICAL PROTEIN, SELENOMET), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Eswaramoorthy, S, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 1999-08-18 | | Release date: | 1999-09-02 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a yeast hypothetical protein selected by a structural genomics approach.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1DP7
 
 | | COCRYSTAL STRUCTURE OF RFX-DBD IN COMPLEX WITH ITS COGNATE X-BOX BINDING SITE | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*CP*GP*(BRU)P*TP*AP*CP*CP*AP*(BRU)P*GP*GP*TP*AP*AP*CP*G)-3'), ... | | Authors: | Gajiwala, K.S, Chen, H, Cornille, F, Roques, B.P, Reith, W, Mach, B, Burley, S.K. | | Deposit date: | 1999-12-23 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding.
Nature, 403, 2000
|
|
