8QQK
 
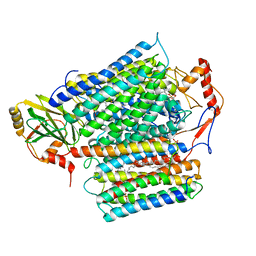 | | Cryo-EM structure of E. coli cytochrome bo3 quinol oxidase assembled in peptidiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, ... | | Authors: | Gao, Y, Zhang, Y, Hakke, S, Peters, P.J, Ravelli, R.B.G. | | Deposit date: | 2023-10-05 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of cytochrome bo 3 quinol oxidase assembled in peptidiscs reveals an "open" conformation for potential ubiquinone-8 release.
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
8ZHU
 
 | |
8ZHV
 
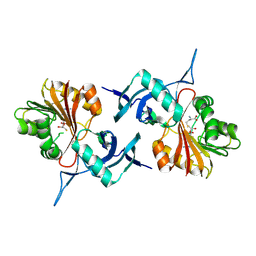 | | Structure of 3-amino-3-carboxyltransferase in complex with SAH and nocardicin G in the biosynthesis of nocardicins | | Descriptor: | 5'-thioadenosine, Isonocardicin synthase, Nocardicin G, ... | | Authors: | Gao, Y, Mori, T, Awakawa, T, Abe, I. | | Deposit date: | 2024-05-11 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure of 3-amino-3-carboxyltransferase in complex with SAH and nocardicin G in the biosynthesis of nocardicins
To Be Published
|
|
5KFY
 
 | |
5KFE
 
 | |
5KFO
 
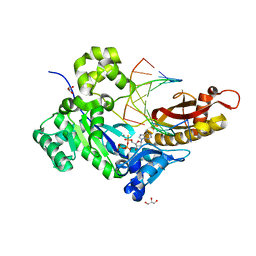 | |
5KG1
 
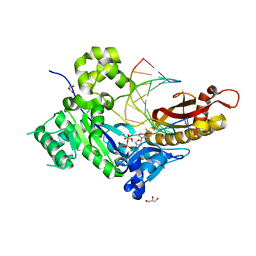 | |
5KFC
 
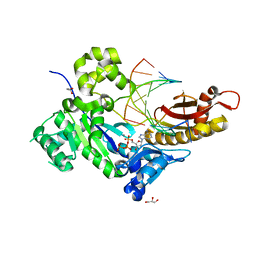 | |
5KFL
 
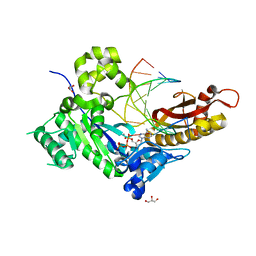 | |
5KFX
 
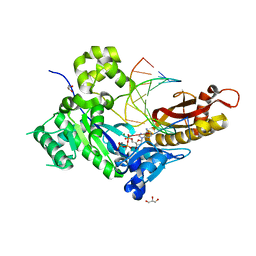 | |
6PZQ
 
 | |
7JSN
 
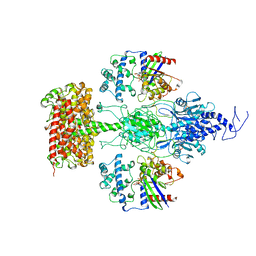 | | Structure of the Visual Signaling Complex between Transducin and Phosphodiesterase 6 | | Descriptor: | 2-{2-ETHOXY-5-[(4-ETHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}-5-METHYL-7-PROPYLIMIDAZO[5,1-F][1,2,4]TRIAZIN-4(1H)-ONE, GUANOSINE-3',5'-MONOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gao, Y, Eskici, G, Ramachandran, S, Skiniotis, G, Cerione, R.A. | | Deposit date: | 2020-08-15 | | Release date: | 2020-10-21 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the Visual Signaling Complex between Transducin and Phosphodiesterase 6.
Mol.Cell, 80, 2020
|
|
6OY9
 
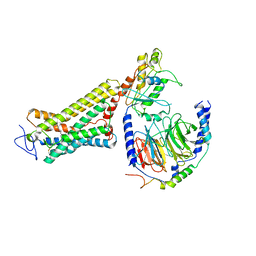 | | Structure of the Rhodopsin-Transducin Complex | | Descriptor: | Gt-alpha/Gi1-alpha chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, ... | | Authors: | Gao, Y, Hu, H, Ramachandran, S, Erickson, J.W, Cerione, R.A, Skiniotis, G. | | Deposit date: | 2019-05-14 | | Release date: | 2019-07-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of the Rhodopsin-Transducin Complex: Insights into G-Protein Activation.
Mol.Cell, 75, 2019
|
|
6OYA
 
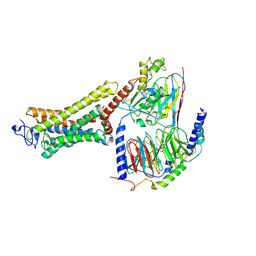 | | Structure of the Rhodopsin-Transducin-Nanobody Complex | | Descriptor: | Camelid antibody VHH fragment, Gt-alpha/Gi1-alpha chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Gao, Y, Hu, H, Ramachandran, S, Erickson, J.W, Cerione, R.A, Skiniotis, G. | | Deposit date: | 2019-05-14 | | Release date: | 2019-07-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of the Rhodopsin-Transducin Complex: Insights into G-Protein Activation.
Mol.Cell, 75, 2019
|
|
5KFG
 
 | |
5KFR
 
 | |
5KG4
 
 | |
5KFK
 
 | |
5KFU
 
 | |
5KG6
 
 | |
5KFA
 
 | |
5KFJ
 
 | |
5KFV
 
 | |
5KG7
 
 | |
5KFP
 
 | |
