5X9Y
 
 | |
5B42
 
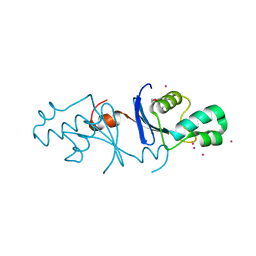 | | Crystal structure of the C-terminal endonuclease domain of Aquifex aeolicus MutL. | | Descriptor: | CADMIUM ION, DI(HYDROXYETHYL)ETHER, DNA mismatch repair protein MutL | | Authors: | Fukui, K, Baba, S, Kumasaka, T, Yano, T. | | Deposit date: | 2016-03-30 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Features and Functional Dependency on beta-Clamp Define Distinct Subfamilies of Bacterial Mismatch Repair Endonuclease MutL
J.Biol.Chem., 291, 2016
|
|
7VUF
 
 | |
7VUK
 
 | |
8H1E
 
 | |
8H1G
 
 | |
8H1F
 
 | |
2ZQE
 
 | | Crystal structure of the Smr domain of Thermus thermophilus MutS2 | | Descriptor: | MutS2 protein | | Authors: | Fukui, K, Kitamura, Y, Nakagawa, N, Masui, R, Kuramitsu, S. | | Deposit date: | 2008-08-08 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of MutS2 endonuclease domain and the mechanism of homologous recombination suppression
J.Biol.Chem., 283, 2008
|
|
5Z41
 
 | | Aquifex aeolicus MutL endonuclease domain with a single zinc ion. | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA mismatch repair protein MutL, MAGNESIUM ION, ... | | Authors: | Fukui, K, Yano, T. | | Deposit date: | 2018-01-10 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Multiple zinc ions maintain the open conformation of the catalytic site in the DNA mismatch repair endonuclease MutL from Aquifex aeolicus
FEBS Lett., 592, 2018
|
|
5Z42
 
 | | Aquifex aeolicus MutL endonuclease domain with three zinc ions. | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, DNA mismatch repair protein MutL, ... | | Authors: | Fukui, K, Yano, T. | | Deposit date: | 2018-01-10 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Multiple zinc ions maintain the open conformation of the catalytic site in the DNA mismatch repair endonuclease MutL from Aquifex aeolicus
FEBS Lett., 592, 2018
|
|
6LZJ
 
 | | Aquifex aeolicus MutL ATPase domain complexed with AMPPCP | | Descriptor: | DNA mismatch repair protein MutL, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Fukui, K, Izuhara, K, Yano, T. | | Deposit date: | 2020-02-19 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.72835565 Å) | | Cite: | A Lynch syndrome-associated mutation at a Bergerat ATP-binding fold destabilizes the structure of the DNA mismatch repair endonuclease MutL.
J.Biol.Chem., 295, 2020
|
|
6LZI
 
 | | Aquifex aeolicus MutL ATPase domain complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, DNA mismatch repair protein MutL, ... | | Authors: | Fukui, K, Izuhara, K, Yano, T. | | Deposit date: | 2020-02-19 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69105685 Å) | | Cite: | A Lynch syndrome-associated mutation at a Bergerat ATP-binding fold destabilizes the structure of the DNA mismatch repair endonuclease MutL.
J.Biol.Chem., 295, 2020
|
|
6LZK
 
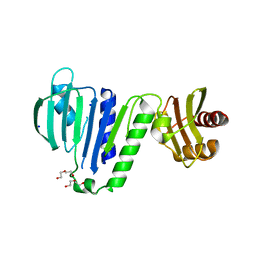 | | Aquifex aeolicus MutL ATPase domain with K252N mutation | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA mismatch repair protein MutL, SODIUM ION | | Authors: | Fukui, K, Izuhara, K, Yano, T. | | Deposit date: | 2020-02-19 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.15949631 Å) | | Cite: | A Lynch syndrome-associated mutation at a Bergerat ATP-binding fold destabilizes the structure of the DNA mismatch repair endonuclease MutL.
J.Biol.Chem., 295, 2020
|
|
4U4P
 
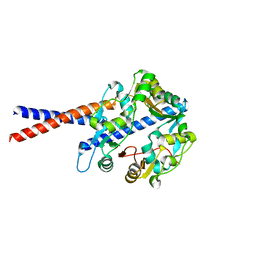 | | Crystal structure of the human condensin SMC hinge domain heterodimer with short coiled coils | | Descriptor: | Structural maintenance of chromosomes protein 2, Structural maintenance of chromosomes protein 4 | | Authors: | Uchiyama, S, Kawahara, K, Hosokawa, Y, Fukakusa, S, Oki, H, Nakamura, S, Noda, M, Takino, R, Miyahara, Y, Maruno, T, Kobayashi, Y, Ohkubo, T, Fukui, K. | | Deposit date: | 2014-07-24 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for dimer information and DNA recognition of human SMC proteins
to be published
|
|
5H45
 
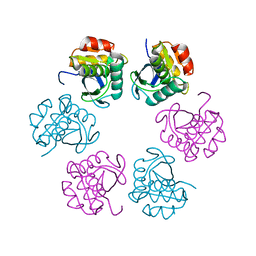 | | Crystal structure of the C-terminal Lon protease-like domain of Thermus thermophilus RadA/Sms | | Descriptor: | DNA repair protein RadA | | Authors: | Inoue, M, Fukui, K, Fujii, Y, Nakagawa, N, Yano, T, Kuramitsu, S, Masui, R. | | Deposit date: | 2016-10-30 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Lon protease-like domain in the bacterial RecA paralog RadA is required for DNA binding and repair.
J. Biol. Chem., 292, 2017
|
|
3A4U
 
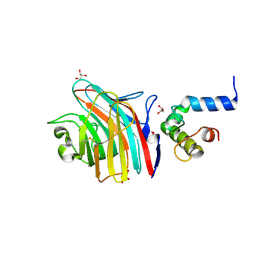 | | Crystal structure of MCFD2 in complex with carbohydrate recognition domain of ERGIC-53 | | Descriptor: | CALCIUM ION, GLYCEROL, Multiple coagulation factor deficiency protein 2, ... | | Authors: | Nishio, M, Kamiya, Y, Mizushima, T, Wakatsuki, S, Sasakawa, H, Yamamoto, K, Uchiyama, S, Noda, M, McKay, A.R, Fukui, K, Hauri, H.P, Kato, K. | | Deposit date: | 2009-07-17 | | Release date: | 2010-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for the cooperative interplay between the two causative gene products of combined factor V and factor VIII deficiency.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2YZL
 
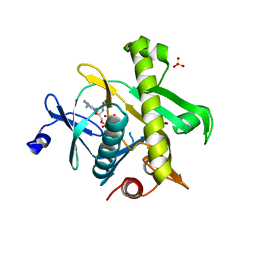 | | Crystal structure of phosphoribosylaminoimidazole-succinocarboxamide synthase with ADP from Methanocaldococcus jannaschii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphoribosylaminoimidazole-succinocarboxamide synthase, SULFATE ION | | Authors: | Kanagawa, M, Baba, S, Fukui, K, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-06 | | Release date: | 2007-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of phosphoribosylaminoimidazole-succinocarboxamide synthase with ADP from Methanocaldococcus jannaschii
To be Published
|
|
5XE9
 
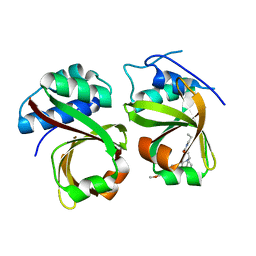 | | Crystal Structure of the Complex of the Peptidase Domain of Streptococcus mutans ComA with a Small Molecule Inhibitor. | | Descriptor: | Putative ABC transporter, ATP-binding protein ComA, [(1~{S},2~{R},4~{S},5~{R})-5-[5-(4-methoxyphenyl)-2-methyl-pyrazol-3-yl]-1-azabicyclo[2.2.2]octan-2-yl]methyl ~{N}-propylcarbamate | | Authors: | Ishii, S, Fukui, K, Yokoshima, S, Kumagai, K, Beniyama, Y, Kodama, T, Fukuyama, T, Okabe, T, Nagano, T, Kojima, H, Yano, T. | | Deposit date: | 2017-04-03 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | High-throughput Screening of Small Molecule Inhibitors of the Streptococcus Quorum-sensing Signal Pathway
Sci Rep, 7, 2017
|
|
5XE8
 
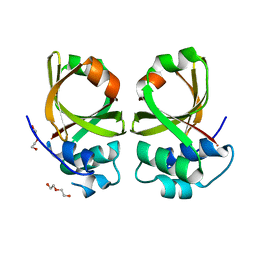 | | Crystal Structure of the Peptidase Domain of Streptococcus mutans ComA | | Descriptor: | DI(HYDROXYETHYL)ETHER, Putative ABC transporter, ATP-binding protein ComA | | Authors: | Ishii, S, Fukui, K, Yokoshima, S, Kumagai, K, Beniyama, Y, Kodama, T, Fukuyama, T, Okabe, T, Nagano, T, Kojima, H, Yano, T. | | Deposit date: | 2017-04-03 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | High-throughput Screening of Small Molecule Inhibitors of the Streptococcus Quorum-sensing Signal Pathway
Sci Rep, 7, 2017
|
|
5YTQ
 
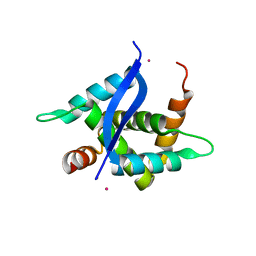 | | Crystal Structure of TTHA0139 L34A with Lanthanum from Thermus thermophilus HB8 | | Descriptor: | LANTHANUM (III) ION, TTHA0139 | | Authors: | Takao, K, Inoue, M, Fukui, K, Yano, T, Masui, R. | | Deposit date: | 2017-11-19 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Crystal Structure of TTHA0139 L34A with Lanthanum from Thermus thermophilus HB8
To Be Published
|
|
5YTP
 
 | |
5ZJ9
 
 | | human D-amino acid oxidase complexed with 5-chlorothiophene-3-carboxylic acid | | Descriptor: | 5-chloro thiophene-3-carboxylic acid, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kato, Y, Hin, N, Maita, N, Thomas, A.G, Kurosawa, S, Rojas, C, Yorita, K, Slusher, B.S, Fukui, K, Tsukamoto, T. | | Deposit date: | 2018-03-19 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for potent inhibition of d-amino acid oxidase by thiophene carboxylic acids
Eur J Med Chem, 159, 2018
|
|
5ZJA
 
 | | human D-amino acid oxidase complexed with 5-chlorothiophene-2-carboxylic acid | | Descriptor: | 5-chloro thiophene-2-carboxylic acid, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kato, Y, Hin, N, Maita, N, Thomas, A.G, Kurosawa, S, Rojas, C, Yorita, K, Slusher, B.S, Fukui, K, Tsukamoto, T. | | Deposit date: | 2018-03-19 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for potent inhibition of d-amino acid oxidase by thiophene carboxylic acids
Eur J Med Chem, 159, 2018
|
|
3WJ4
 
 | | Crystal structure of PPARgamma ligand binding domain in complex with tributyltin | | Descriptor: | Peroxisome proliferator-activated receptor gamma, tributylstannanyl | | Authors: | Harada, S, Hiromori, Y, Fukakusa, S, Kawahara, K, Nakamura, S, Noda, M, Uchiyama, S, Fukui, K, Nishikawa, J, Nagase, H, Kobayashi, Y, Ohkubo, T, Yoshida, T, Nakanishi, T. | | Deposit date: | 2013-10-04 | | Release date: | 2014-10-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for PPARgamma transactivation by endocrine disrupting organotin compounds
To be Published
|
|
3WJ5
 
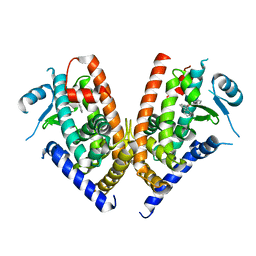 | | Crystal structure of PPARgamma ligand binding domain in complex with triphenyltin | | Descriptor: | Peroxisome proliferator-activated receptor gamma, triphenylstannanyl | | Authors: | Harada, S, Hiromori, Y, Fukakusa, S, Kawahara, K, Nakamura, S, Noda, M, Uchiyama, S, Fukui, K, Nishikawa, J, Nagase, H, Kobayashi, Y, Ohkubo, T, Yoshida, T, Nakanishi, T. | | Deposit date: | 2013-10-04 | | Release date: | 2014-10-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for PPARgamma transactivation by endocrine disrupting organotin compounds
To be Published
|
|
