6ADH
 
 | |
1ABA
 
 | | THE STRUCTURE OF OXIDIZED BACTERIOPHAGE T4 GLUTAREDOXIN (THIOREDOXIN). REFINEMENT OF NATIVE AND MUTANT PROTEINS | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTAREDOXIN | | Authors: | Eklund, H, Ingelman, M, Soderberg, B.-O, Uhlin, T, Nordlund, P, Nikkola, M, Sonnerstam, U, Joelson, T, Petratos, K. | | Deposit date: | 1992-04-24 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of oxidized bacteriophage T4 glutaredoxin (thioredoxin). Refinement of native and mutant proteins.
J.Mol.Biol., 228, 1992
|
|
1AAZ
 
 | | THE STRUCTURE OF OXIDIZED BACTERIOPHAGE T4 GLUTAREDOXIN (THIOREDOXIN) | | Descriptor: | CADMIUM ION, GLUTAREDOXIN | | Authors: | Eklund, H, Ingelman, M, Soderberg, B.-O, Uhlin, T, Nordlund, P, Nikkola, M, Sonnerstam, U, Joelson, T, Petratos, K. | | Deposit date: | 1992-04-24 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of oxidized bacteriophage T4 glutaredoxin (thioredoxin). Refinement of native and mutant proteins.
J.Mol.Biol., 228, 1992
|
|
5ADH
 
 | |
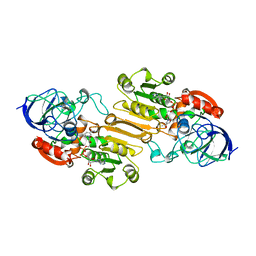
1CDO
 
 | |
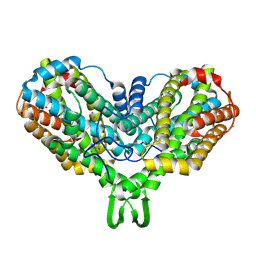
1MRR
 
 | |
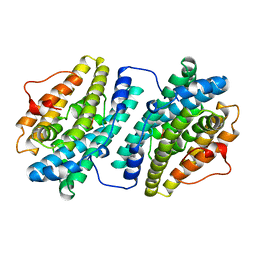
2R2F
 
 | |
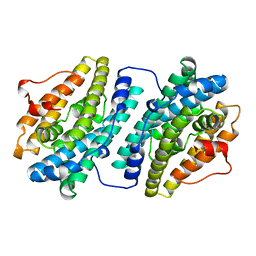
1R2F
 
 | |
1RIB
 
 | |
7ADH
 
 | |
8ADH
 
 | |
4R1R
 
 | |
7R1R
 
 | | RIBONUCLEOTIDE REDUCTASE E441Q MUTANT R1 PROTEIN FROM ESCHERICHIA COLI | | Descriptor: | RIBONUCLEOTIDE REDUCTASE R1 PROTEIN, RIBONUCLEOTIDE REDUCTASE R2 PROTEIN | | Authors: | Eriksson, M, Eklund, H. | | Deposit date: | 1997-09-17 | | Release date: | 1998-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A new mechanism-based radical intermediate in a mutant R1 protein affecting the catalytically essential Glu441 in Escherichia coli ribonucleotide reductase.
J.Biol.Chem., 272, 1997
|
|
5R1R
 
 | | RIBONUCLEOTIDE REDUCTASE E441A MUTANT R1 PROTEIN FROM ESCHERICHIA COLI | | Descriptor: | RIBONUCLEOTIDE REDUCTASE R1 PROTEIN, RIBONUCLEOTIDE REDUCTASE R2 PROTEIN | | Authors: | Eriksson, M, Eklund, H. | | Deposit date: | 1997-09-17 | | Release date: | 1998-03-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A new mechanism-based radical intermediate in a mutant R1 protein affecting the catalytically essential Glu441 in Escherichia coli ribonucleotide reductase.
J.Biol.Chem., 272, 1997
|
|
6R1R
 
 | | RIBONUCLEOTIDE REDUCTASE E441D MUTANT R1 PROTEIN FROM ESCHERICHIA COLI | | Descriptor: | RIBONUCLEOTIDE REDUCTASE R1 PROTEIN, RIBONUCLEOTIDE REDUCTASE R2 PROTEIN | | Authors: | Eriksson, M, Eklund, H. | | Deposit date: | 1997-09-17 | | Release date: | 1998-03-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A new mechanism-based radical intermediate in a mutant R1 protein affecting the catalytically essential Glu441 in Escherichia coli ribonucleotide reductase.
J.Biol.Chem., 272, 1997
|
|
3R1R
 
 | |
7MDH
 
 | | STRUCTURAL BASIS FOR LIGHT ACITVATION OF A CHLOROPLAST ENZYME. THE STRUCTURE OF SORGHUM NADP-MALATE DEHYDROGENASE IN ITS OXIDIZED FORM | | Descriptor: | PROTEIN (MALATE DEHYDROGENASE), ZINC ION | | Authors: | Johansson, K, Ramaswamy, S, Saarinen, M, Lemaire-Chamley, M, Issakidis-Bourguet, E, Miginiac-Maslow, M, Eklund, H. | | Deposit date: | 1999-02-16 | | Release date: | 1999-06-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for light activation of a chloroplast enzyme: the structure of sorghum NADP-malate dehydrogenase in its oxidized form.
Biochemistry, 38, 1999
|
|
1HLD
 
 | | STRUCTURES OF HORSE LIVER ALCOHOL DEHYDROGENASE COMPLEXED WITH NAD+ AND SUBSTITUTED BENZYL ALCOHOLS | | Descriptor: | 2,3,4,5,6-PENTAFLUOROBENZYL ALCOHOL, ALCOHOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Ramaswamy, S, Eklund, H, Plapp, B.V. | | Deposit date: | 1993-11-23 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of horse liver alcohol dehydrogenase complexed with NAD+ and substituted benzyl alcohols.
Biochemistry, 33, 1994
|
|
1GV0
 
 | | Structural Basis for Thermophilic Protein Stability: Structures of Thermophilic and Mesophilic Malate Dehydrogenases | | Descriptor: | MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dalhus, B, Sarinen, M, Sauer, U.H, Eklund, P, Johansson, K, Karlsson, A, Ramaswamy, S, Bjork, A, Synstad, B, Naterstad, K, Sirevag, R, Eklund, H. | | Deposit date: | 2002-02-04 | | Release date: | 2002-02-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Thermophilic Protein Stability: Structures of Thermophilic and Mesophilic Malate Dehydrogenases
J.Mol.Biol., 318, 2002
|
|
1GUY
 
 | | Structural Basis for Thermophilic Protein Stability: Structures of Thermophilic and Mesophilic Malate Dehydrogenases | | Descriptor: | CADMIUM ION, MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dalhus, B, Sarinen, M, Sauer, U.H, Eklund, P, Johansson, K, Karlsson, A, Ramaswamy, S, Bjork, A, Synstad, B, Naterstad, K, Sirevag, R, Eklund, H. | | Deposit date: | 2002-02-04 | | Release date: | 2002-02-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Thermophilic Protein Stability: Structures of Thermophilic and Mesophilic Malate Dehydrogenases
J.Mol.Biol., 318, 2002
|
|
1GUZ
 
 | | Structural Basis for Thermophilic Protein Stability: Structures of Thermophilic and Mesophilic Malate Dehydrogenases | | Descriptor: | MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dalhus, B, Sarinen, M, Sauer, U.H, Eklund, P, Johansson, K, Karlsson, A, Ramaswamy, S, Bjork, A, Synstad, B, Naterstad, K, Sirevag, R, Eklund, H. | | Deposit date: | 2002-02-04 | | Release date: | 2002-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Thermophilic Protein Stability: Structures of Thermophilic and Mesophilic Malate Dehydrogenases
J.Mol.Biol., 318, 2002
|
|
1GV1
 
 | | Structural Basis for Thermophilic Protein Stability: Structures of Thermophilic and Mesophilic Malate Dehydrogenases | | Descriptor: | MALATE DEHYDROGENASE | | Authors: | Dalhus, B, Sarinen, M, Sauer, U.H, Eklund, P, Johansson, K, Karlsson, A, Ramaswamy, S, Bjork, A, Synstad, B, Naterstad, K, Sirevag, R, Eklund, H. | | Deposit date: | 2002-02-04 | | Release date: | 2002-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Thermophilic Protein Stability: Structures of Thermophilic and Mesophilic Malate Dehydrogenases
J.Mol.Biol., 318, 2002
|
|
1XIK
 
 | | RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 BETA CHAIN | | Descriptor: | FE (II) ION, MERCURY (II) ION, PROTEIN R2 OF RIBONUCLEOTIDE REDUCTASE | | Authors: | Logan, D.T, Su, X.-D, Aberg, A, Regnstrom, K, Hajdu, J, Eklund, H, Nordlund, P. | | Deposit date: | 1996-08-06 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of reduced protein R2 of ribonucleotide reductase: the structural basis for oxygen activation at a dinuclear iron site.
Structure, 4, 1996
|
|
1XSM
 
 | | PROTEIN R2 OF RIBONUCLEOTIDE REDUCTASE FROM MOUSE | | Descriptor: | FE (III) ION, RIBONUCLEOTIDE REDUCTASE R2 | | Authors: | Kauppi, B, Nielsen, B.N, Ramaswamy, S, Kjoller-Larsen, I, Thelander, M, Thelander, L, Eklund, H. | | Deposit date: | 1996-07-03 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The three-dimensional structure of mammalian ribonucleotide reductase protein R2 reveals a more-accessible iron-radical site than Escherichia coli R2.
J.Mol.Biol., 262, 1996
|
|
3DQY
 
 | | Crystal structure of Toluene 2,3-Dioxygenase Ferredoxin | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Toluene 1,2-dioxygenase system ferredoxin subunit | | Authors: | Friemann, R, Lee, K, Brown, E.N, Gibson, D.T, Eklund, H, Ramaswamy, S. | | Deposit date: | 2008-07-10 | | Release date: | 2009-03-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structures of the multicomponent Rieske non-heme iron toluene 2,3-dioxygenase enzyme system
Acta Crystallogr.,Sect.D, 65, 2009
|
|
