2XB2
 
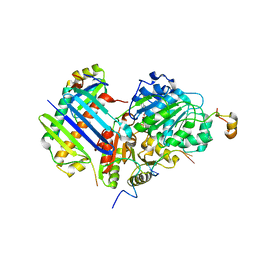 | | Crystal structure of the core Mago-Y14-eIF4AIII-Barentsz-UPF3b assembly shows how the EJC is bridged to the NMD machinery | | Descriptor: | EUKARYOTIC INITIATION FACTOR 4A-III, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Buchwald, G, Ebert, J, Basquin, C, Sauliere, J, Jayachandran, U, Bono, F, Le Hir, H, Conti, E. | | Deposit date: | 2010-04-03 | | Release date: | 2010-05-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Insights Into the Recruitment of the Nmd Machinery from the Crystal Structure of a Core Ejc-Upf3B Complex.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4W8Z
 
 | |
4W8W
 
 | |
1RK8
 
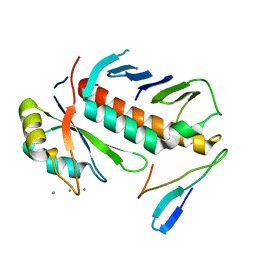 | | Structure of the cytosolic protein PYM bound to the Mago-Y14 core of the exon junction complex | | Descriptor: | CALCIUM ION, CG8781-PA protein, Mago nashi protein, ... | | Authors: | Bono, F, Ebert, J, Guettler, T, Izaurralde, E, Conti, E. | | Deposit date: | 2003-11-21 | | Release date: | 2004-04-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular insights into the interaction of PYM with
the Mago-Y14 core of the exon junction complex
EMBO Rep., 5, 2004
|
|
4W8V
 
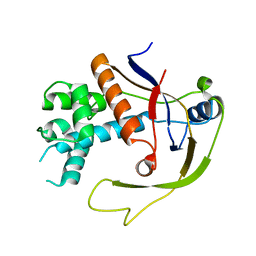 | |
4W8X
 
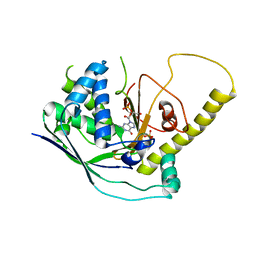 | | Crystal Structure of Cmr1 from Pyrococcus furiosus bound to a nucleotide | | Descriptor: | CRISPR system Cmr subunit Cmr1-1, GUANOSINE-3'-MONOPHOSPHATE, PHOSPHATE ION | | Authors: | Benda, C, Ebert, J, Baumgaertner, M, Conti, E. | | Deposit date: | 2014-08-26 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Model of a CRISPR RNA-Silencing Complex Reveals the RNA-Target Cleavage Activity in Cmr4.
Mol.Cell, 56, 2014
|
|
4W8Y
 
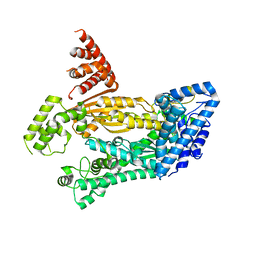 | | Structure of full length Cmr2 from Pyrococcus furiosus (Manganese bound form) | | Descriptor: | CRISPR system Cmr subunit Cmr2, MANGANESE (II) ION, ZINC ION | | Authors: | Benda, C, Ebert, J, Baumgaertner, M, Conti, E. | | Deposit date: | 2014-08-26 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Model of a CRISPR RNA-Silencing Complex Reveals the RNA-Target Cleavage Activity in Cmr4.
Mol.Cell, 56, 2014
|
|
4N0L
 
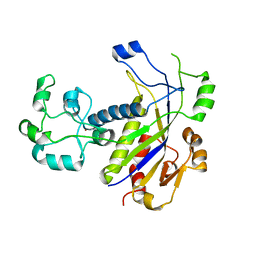 | | Methanopyrus kandleri Csm3 crystal structure | | Descriptor: | Predicted component of a thermophile-specific DNA repair system, contains a RAMP domain, ZINC ION | | Authors: | Hrle, A, Su, A.A, Ebert, J, Benda, C, Conti, E, Randau, L. | | Deposit date: | 2013-10-02 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure and RNA-binding properties of the Type III-A CRISPR-associated protein Csm3.
Rna Biol., 10, 2013
|
|
1Z3H
 
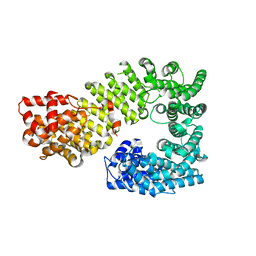 | | The exportin Cse1 in its cargo-free, cytoplasmic state | | Descriptor: | Importin alpha re-exporter, MAGNESIUM ION | | Authors: | Cook, A, Fernandez, E, Lindner, D, Ebert, J, Schlenstedt, G, Conti, E. | | Deposit date: | 2005-03-12 | | Release date: | 2005-05-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structure of the nuclear export receptor cse1 in its cytosolic state reveals a closed conformation incompatible with cargo binding
Mol.Cell, 18, 2005
|
|
5JEA
 
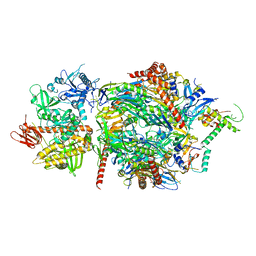 | | Structure of a cytoplasmic 11-subunit RNA exosome complex including Ski7, bound to RNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Exosome complex component CSL4, Exosome complex component MTR3, ... | | Authors: | Kowalinski, E, Ebert, J, Stegmann, E, Conti, E. | | Deposit date: | 2016-04-18 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of a Cytoplasmic 11-Subunit RNA Exosome Complex.
Mol.Cell, 63, 2016
|
|
2WP8
 
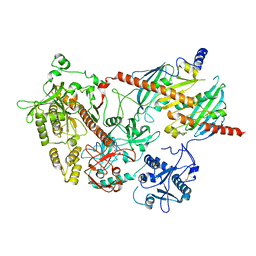 | | yeast rrp44 nuclease | | Descriptor: | CHLORIDE ION, EXOSOME COMPLEX COMPONENT RRP45, EXOSOME COMPLEX COMPONENT SKI6, ... | | Authors: | Basquin, J, Bonneau, F, Ebert, J, Lorentzen, E, Conti, E. | | Deposit date: | 2009-08-03 | | Release date: | 2009-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Yeast Exosome Functions as a Macromolecular Cage to Channel RNA Substrates for Degradation.
Cell(Cambridge,Mass.), 139, 2009
|
|
1YA0
 
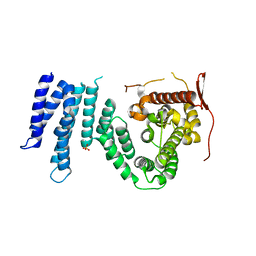 | | Crystal structure of the N-terminal domain of human SMG7 | | Descriptor: | SMG-7 transcript variant 2, SULFATE ION | | Authors: | Fukuhara, N, Ebert, J, Unterholzner, L, Lindner, D, Izaurralde, E, Conti, E. | | Deposit date: | 2004-12-17 | | Release date: | 2005-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | SMG7 Is a 14-3-3-like Adaptor in the Nonsense-Mediated mRNA Decay Pathway.
Mol.Cell, 17, 2005
|
|
4A6Q
 
 | | Crystal structure of mouse SAP18 residues 6-143 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, HISTONE DEACETYLASE COMPLEX SUBUNIT SAP18, ISOPROPYL ALCOHOL | | Authors: | Murachelli, A.G, Ebert, J, Basquin, C, Le Hir, H, Conti, E. | | Deposit date: | 2011-11-08 | | Release date: | 2012-03-07 | | Last modified: | 2015-03-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structure of the Asap Core Complex Reveals the Existence of a Pinin-Containing Psap Complex
Nat.Struct.Mol.Biol., 19, 2012
|
|
4A8X
 
 | | Structure of the core ASAP complex | | Descriptor: | HISTONE DEACETYLASE COMPLEX SUBUNIT SAP18, HOOK-LIKE, ISOFORM A, ... | | Authors: | Murachelli, A.G, Ebert, J, Basquin, C, Le Hir, H, Conti, E. | | Deposit date: | 2011-11-21 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure of the Asap Core Complex Reveals the Existence of a Pinin-Containing Psap Complex
Nat.Struct.Mol.Biol., 19, 2012
|
|
4A90
 
 | | Crystal structure of mouse SAP18 residues 1-143 | | Descriptor: | GLYCEROL, HISTONE DEACETYLASE COMPLEX SUBUNIT SAP18 | | Authors: | Murachelli, A.G, Ebert, J, Basquin, C, Le Hir, H, Conti, E. | | Deposit date: | 2011-11-22 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure of the Asap Core Complex Reveals the Existence of a Pinin-Containing Psap Complex
Nat.Struct.Mol.Biol., 19, 2012
|
|
2QFA
 
 | | Crystal structure of a Survivin-Borealin-INCENP core complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Baculoviral IAP repeat-containing protein 5, Borealin, ... | | Authors: | Jeyaprakash, A.A, Klein, U.R, Lindner, D, Ebert, J, Nigg, E.A, Conti, E. | | Deposit date: | 2007-06-27 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of a Survivin-Borealin-INCENP Core Complex Reveals How Chromosomal Passengers Travel Together.
Cell(Cambridge,Mass.), 131, 2007
|
|
2X1G
 
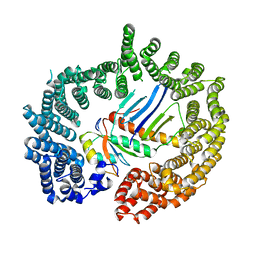 | | Crystal structure of Importin13 - Mago-Y14 complex | | Descriptor: | CADMUS, PROTEIN MAGO NASHI, RNA-BINDING PROTEIN 8A | | Authors: | Bono, F, Cook, A.G, Gruenwald, M, Ebert, J, Conti, E. | | Deposit date: | 2009-12-23 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Nuclear Import Mechanism of the Ejc Component Mago- Y14 Revealed by Structural Studies of Importin 13.
Mol.Cell, 37, 2010
|
|
2X19
 
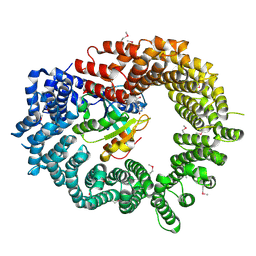 | | Crystal structure of Importin13 - RanGTP complex | | Descriptor: | GTP-BINDING NUCLEAR PROTEIN GSP1/CNR1, GUANOSINE-5'-TRIPHOSPHATE, IMPORTIN-13, ... | | Authors: | Bono, F, Cook, A.G, Gruenwald, M, Ebert, J, Conti, E. | | Deposit date: | 2009-12-23 | | Release date: | 2010-02-16 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nuclear Import Mechanism of the Ejc Component Mago- Y14 Revealed by Structural Studies of Importin 13.
Mol.Cell, 37, 2010
|
|
2J0S
 
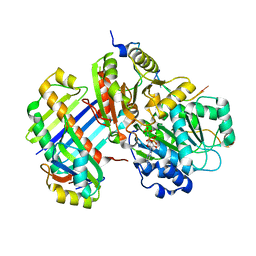 | | The crystal structure of the Exon Junction Complex at 2.2 A resolution | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP *UP*UP*UP*UP*U)-3', ATP-DEPENDENT RNA HELICASE DDX48, MAGNESIUM ION, ... | | Authors: | Bono, F, Ebert, J, Lorentzen, E, Conti, E. | | Deposit date: | 2006-08-04 | | Release date: | 2006-09-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The Crystal Structure of the Exon Junction Complex Reveals How It Mantains a Stable Grip on Mrna
Cell(Cambridge,Mass.), 126, 2006
|
|
2J0Q
 
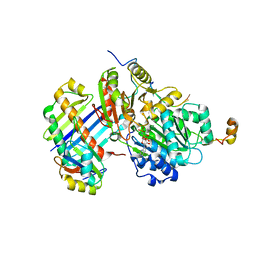 | | The crystal structure of the Exon Junction Complex at 3.2 A resolution | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3', ATP-DEPENDENT RNA HELICASE DDX48, MAGNESIUM ION, ... | | Authors: | Bono, F, Ebert, J, Lorentzen, E, Conti, E. | | Deposit date: | 2006-08-04 | | Release date: | 2006-08-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Crystal Structure of the Exon Junction Complex Reveals How It Maintains a Stable Grip on Mrna.
Cell(Cambridge,Mass.), 126, 2006
|
|
2J0U
 
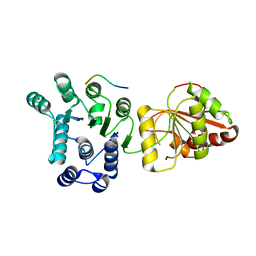 | | The crystal structure of eIF4AIII-Barentsz complex at 3.0 A resolution | | Descriptor: | ATP-DEPENDENT RNA HELICASE DDX48, PROTEIN CASC3 | | Authors: | Bono, F, Ebert, J, Lorentzen, E, Conti, E. | | Deposit date: | 2006-08-04 | | Release date: | 2006-09-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Crystal Structure of the Exon Junction Complex Reveals How It Mantains a Stable Grip on Mrna
Cell(Cambridge,Mass.), 126, 2006
|
|
5OKZ
 
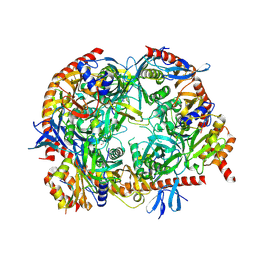 | | Crystal Strucrure of the Mpp6 Exosome complex | | Descriptor: | CHLORIDE ION, Exosome complex component CSL4, Exosome complex component MTR3, ... | | Authors: | Falk, S, Ebert, J, Conti, E. | | Deposit date: | 2017-07-26 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.200039 Å) | | Cite: | Mpp6 Incorporation in the Nuclear Exosome Contributes to RNA Channeling through the Mtr4 Helicase.
Cell Rep, 20, 2017
|
|
4TXD
 
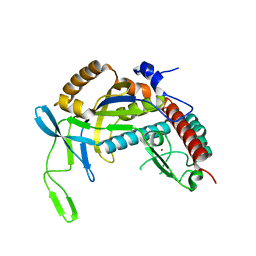 | | Crystal structure of Thermofilum pendens Csc2 | | Descriptor: | Csc2, ZINC ION | | Authors: | Hrle, A, Conti, E. | | Deposit date: | 2014-07-03 | | Release date: | 2014-08-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.818 Å) | | Cite: | Structural analyses of the CRISPR protein Csc2 reveal the RNA-binding interface of the type I-D Cas7 family.
Rna Biol., 11, 2014
|
|
