2W1B
 
 | | The structure of the efflux pump AcrB in complex with bile acid | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, ACRIFLAVIN RESISTANCE PROTEIN B | | Authors: | Drew, D, Klepsch, M.M, Newstead, S, Flaig, R, De Gier, J.W, Iwata, S, Beis, K. | | Deposit date: | 2008-10-17 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | The Structure of the Efflux Pump Acrb in Complex with Bile Acid.
Mol.Membr.Biol., 25, 2008
|
|
7S24
 
 | | Crystal structure of the Na+/H+ antiporter NhaA at pH 6.5 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Na(+)/H(+) antiporter NhaA, PENTAETHYLENE GLYCOL | | Authors: | Drew, D, Brock, J, Uzdavinys, P, Matsuoka, R. | | Deposit date: | 2021-09-03 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Na + /H + antiporter NhaA at active pH reveals the mechanistic basis for pH sensing.
Nat Commun, 13, 2022
|
|
4ATV
 
 | | STRUCTURE OF A TRIPLE MUTANT OF THE NHAA DIMER, CRYSTALLISED AT LOW PH | | Descriptor: | DODECYL-ALPHA-D-MALTOSIDE, NA(+)/H(+) ANTIPORTER NHAA, SULFATE ION | | Authors: | Drew, D, Lee, C, Iwata, S, Cameron, A.D. | | Deposit date: | 2012-05-10 | | Release date: | 2013-07-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the sodium-proton antiporter NhaA dimer and new mechanistic insights.
J. Gen. Physiol., 144, 2014
|
|
4AU5
 
 | | Structure of the NhaA dimer, crystallised at low pH | | Descriptor: | DODECYL-ALPHA-D-MALTOSIDE, NA(+)/H(+) ANTIPORTER NHAA, SULFATE ION | | Authors: | Drew, D, Lee, C, Iwata, S, Cameron, A.D. | | Deposit date: | 2012-05-14 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.696 Å) | | Cite: | Crystal structure of the sodium-proton antiporter NhaA dimer and new mechanistic insights.
J. Gen. Physiol., 144, 2014
|
|
6Z3Y
 
 | |
8PVR
 
 | | Cryo-EM structure of horse Nhe9 bound to PI(3,5)P2 | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,4,6-trihydroxy-3,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, Sodium/hydrogen exchanger 9 | | Authors: | Kokane, S, Meier, P, Gulati, A, Delemotte, L, Drew, D. | | Deposit date: | 2023-07-18 | | Release date: | 2024-07-24 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | PIP 2 -mediated oligomerization of the endosomal sodium/proton exchanger NHE9.
Nat Commun, 16, 2025
|
|
8PXB
 
 | |
7P1K
 
 | | Cryo EM structure of bison NHA2 in nano disc structure | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Phosphatidylinositol, mitochondrial sodium/hydrogen exchanger 9B2 | | Authors: | Matsuoka, R, Fudim, R, Jung, S, Drew, D. | | Deposit date: | 2021-07-01 | | Release date: | 2022-01-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structure, mechanism and lipid-mediated remodeling of the mammalian Na + /H + exchanger NHA2.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P1J
 
 | | Cryo EM structure of bison NHA2 in detergent structure | | Descriptor: | mitochondrial sodium/hydrogen exchanger 9B2 | | Authors: | Matsuoka, R, Fudim, R, Jung, S, Drew, D. | | Deposit date: | 2021-07-01 | | Release date: | 2022-01-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structure, mechanism and lipid-mediated remodeling of the mammalian Na + /H + exchanger NHA2.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P1I
 
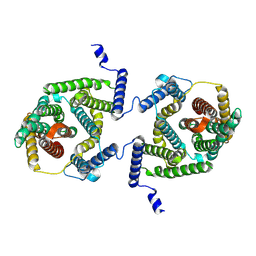 | | Cryo EM structure of bison NHA2 in detergent and N-terminal extension helix | | Descriptor: | mitochondrial sodium/hydrogen exchanger 9B2 | | Authors: | Matsuoka, R, Fudim, R, Jung, S, Drew, D. | | Deposit date: | 2021-07-01 | | Release date: | 2022-01-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structure, mechanism and lipid-mediated remodeling of the mammalian Na + /H + exchanger NHA2.
Nat.Struct.Mol.Biol., 29, 2022
|
|
9QUW
 
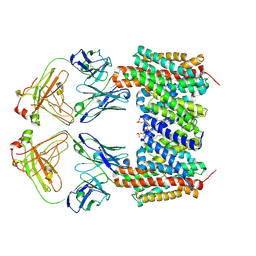 | | Cryo-EM structure of the human NHA2-Fab complex bound to phloretin | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, 3-(4-HYDROXYPHENYL)-1-(2,4,6-TRIHYDROXYPHENYL)PROPAN-1-ONE, Fab heavy chain, ... | | Authors: | Jung, S, Drew, D. | | Deposit date: | 2025-04-11 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and Inhibition of the Human Na + /H + Exchanger SLC9B2.
Int J Mol Sci, 26, 2025
|
|
9QUB
 
 | | Cryo-EM structure of the human NHA2-Fab complex | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, Fab-heavy chain, Fab-light chain, ... | | Authors: | Jung, S, Drew, D. | | Deposit date: | 2025-04-10 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure and Inhibition of the Human Na + /H + Exchanger SLC9B2.
Int J Mol Sci, 26, 2025
|
|
8PS0
 
 | |
8OTW
 
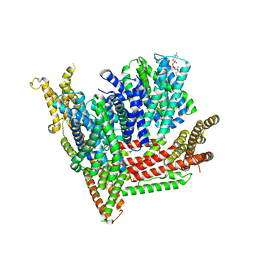 | | Cryo-EM structure of Strongylocentrotus purpuratus SLC9C1 in presence of cAMP | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Sperm-specific sodium proton exchanger | | Authors: | Yeo, H, Mehta, V, Gulati, A, Drew, D. | | Deposit date: | 2023-04-21 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structure and electromechanical coupling of a voltage-gated Na + /H + exchanger.
Nature, 623, 2023
|
|
4D1C
 
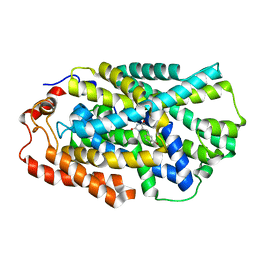 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH bromovinylhydantoin bound. | | Descriptor: | (5Z)-5-[(3-bromophenyl)methylidene]imidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | Authors: | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
4D1D
 
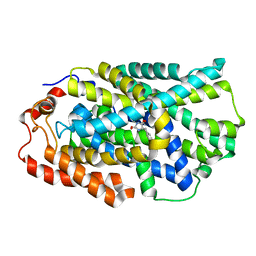 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER with the inhibitor 5-(2-naphthylmethyl)-L-hydantoin. | | Descriptor: | 5-(2-NAPHTHYLMETHYL)-D-HYDANTOIN, 5-(2-NAPHTHYLMETHYL)-L-HYDANTOIN, HYDANTOIN TRANSPORT PROTEIN, ... | | Authors: | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
4D1B
 
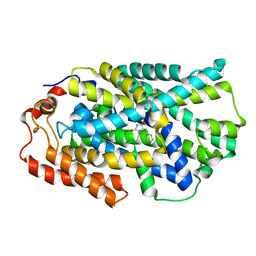 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH BENZYL-HYDANTOIN | | Descriptor: | (5S)-5-benzylimidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | Authors: | Brueckner, F, Geng, T, Weyand, S, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
4D1A
 
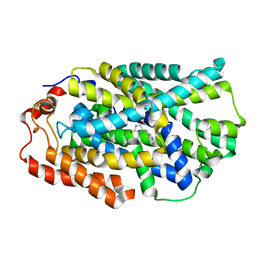 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH INDOLYLMETHYL-HYDANTOIN | | Descriptor: | (5S)-5-(1H-indol-3-ylmethyl)imidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | Authors: | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
8OTX
 
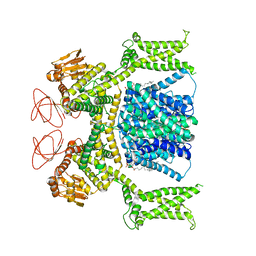 | | Cryo-EM structure of Strongylocentrotus purpuratus sperm-specific Na+/H+ exchanger SLC9C1 in nanodisc | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, Sperm-specific sodium proton exchanger | | Authors: | Yeo, H, Mehta, V, Gulati, A, Drew, D. | | Deposit date: | 2023-04-21 | | Release date: | 2023-11-01 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structure and electromechanical coupling of a voltage-gated Na + /H + exchanger.
Nature, 623, 2023
|
|
8OTQ
 
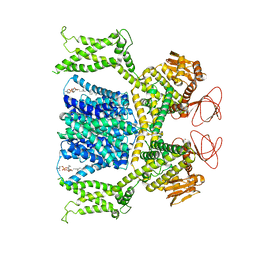 | | Cryo-EM structure of Strongylocentrotus purpuratus sperm-specific Na+/H+ exchanger SLC9C1 in GDN | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, PALMITIC ACID, Sperm-specific sodium proton exchanger | | Authors: | Yeo, H, Mehta, V, Gulati, A, Drew, D. | | Deposit date: | 2023-04-21 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structure and electromechanical coupling of a voltage-gated Na + /H + exchanger.
Nature, 623, 2023
|
|
9EMB
 
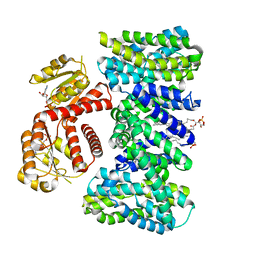 | | Structure of KefC Asp156Asn variant | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, ADENOSINE MONOPHOSPHATE, Glutathione-regulated potassium-efflux system protein KefC | | Authors: | Gulati, A, Kokane, S, Drew, D. | | Deposit date: | 2024-03-07 | | Release date: | 2024-06-05 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structure and mechanism of the K + /H + exchanger KefC.
Nat Commun, 15, 2024
|
|
6Z3Z
 
 | |
9GSL
 
 | |
9GRY
 
 | |
9I20
 
 | | Cryo-EM structure of human SLC35B1 with ADP | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, ADENOSINE-5'-DIPHOSPHATE, Maltodextrin-binding protein, ... | | Authors: | Gulati, A, Ahn, D, Suades, A, Drew, D. | | Deposit date: | 2025-01-17 | | Release date: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structure and step-wise nucleotide translocation of the human ATP/ADP exchanger SLC35B1
To Be Published
|
|
