3B63
 
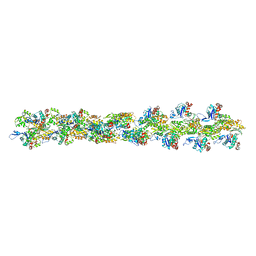 | | Actin filament model in the extended form of acromsomal bundle in the Limulus sperm | | Descriptor: | Actin | | Authors: | Cong, Y, Topf, M, Sali, A, Matsudaira, P, Dougherty, M, Chiu, W, Schmid, M.F. | | Deposit date: | 2007-10-26 | | Release date: | 2008-11-18 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Crystallographic conformers of actin in a biologically active bundle of filaments.
J.Mol.Biol., 375, 2008
|
|
3B5U
 
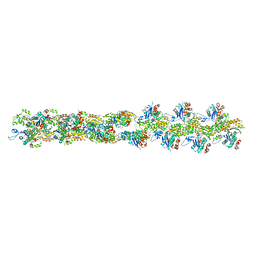 | | Actin filament model from extended form of acromsomal bundle in the Limulus sperm | | Descriptor: | Actin, alpha skeletal muscle | | Authors: | Cong, Y, Topf, M, Sali, A, Matsudaira, P, Dougherty, M, Chiu, W, Schmid, M.F. | | Deposit date: | 2007-10-26 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | ELECTRON CRYSTALLOGRAPHY (9.5 Å) | | Cite: | Crystallographic conformers of actin in a biologically active bundle of filaments
J.Mol.Biol., 375, 2008
|
|
2XD8
 
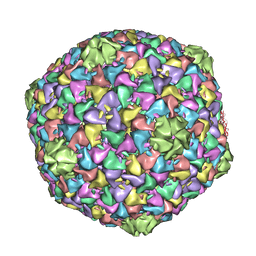 | | Capsid structure of the infectious Prochlorococcus Cyanophage P-SSP7 | | Descriptor: | T7-LIKE CAPSID PROTEIN | | Authors: | Liu, X, Zhang, Q, Murata, K, Baker, M.L, Sullivan, M.B, Fu, C, Dougherty, M, Schmid, M.F, Osburne, M.S, Chisholm, S.W, Chiu, W. | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural Changes in a Marine Podovirus Associated with Release of its Genome Into Prochlorococcus
Nat.Struct.Mol.Biol., 17, 2010
|
|
2XYZ
 
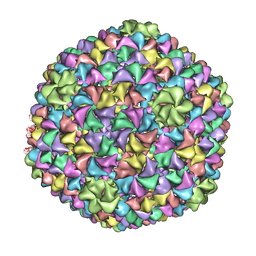 | | De Novo model of Bacteriophage P22 virion coat protein | | Descriptor: | COAT PROTEIN | | Authors: | Chen, D.-H, Baker, M.L, Hryc, C.F, DiMaio, F, Jakana, J, Wu, W, Dougherty, M, Haase-Pettingell, C, Schmid, M.F, Jiang, W, Baker, D, King, J.A, Chiu, W. | | Deposit date: | 2010-11-19 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural Basis for Scaffolding-Mediated Assembly and Maturation of a DsDNA Virus.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2XYY
 
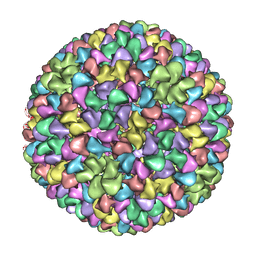 | | De Novo model of Bacteriophage P22 procapsid coat protein | | Descriptor: | COAT PROTEIN | | Authors: | Chen, D.-H, Baker, M.L, Hryc, C.F, DiMaio, F, Jakana, J, Wu, W, Dougherty, M, Haase-Pettingell, C, Schmid, M.F, Jiang, W, Baker, D, King, J.A, Chiu, W. | | Deposit date: | 2010-11-19 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Basis for Scaffolding-Mediated Assembly and Maturation of a DsDNA Virus.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3IYF
 
 | | Atomic Model of the Lidless Mm-cpn in the Open State | | Descriptor: | Chaperonin | | Authors: | Zhang, J, Baker, M.L, Schroeder, G, Douglas, N.R, Reissmann, S, Jakana, J, Dougherty, M, Fu, C.J, Levitt, M, Ludtke, S.J, Frydman, J, Chiu, W. | | Deposit date: | 2009-10-23 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Mechanism of folding chamber closure in a group II chaperonin
Nature, 463, 2010
|
|
3LOS
 
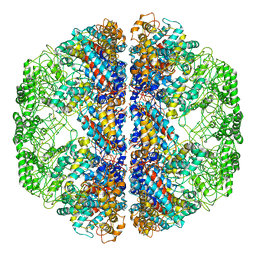 | | Atomic Model of Mm-cpn in the Closed State | | Descriptor: | Chaperonin | | Authors: | Zhang, J, Baker, M.L, Schroeder, G, Douglas, N.R, Reissmann, S, Jakana, J, Dougherty, M, Fu, C.J, Levitt, M, Ludtke, S.J, Frydman, J, Chiu, W. | | Deposit date: | 2010-02-04 | | Release date: | 2010-03-16 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mechanism of folding chamber closure in a group II chaperonin
Nature, 463, 2010
|
|
1TZ3
 
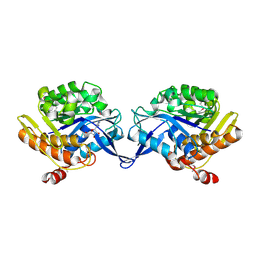 | | crystal structure of aminoimidazole riboside kinase complexed with aminoimidazole riboside | | Descriptor: | 5-AMINOIMIDAZOLE RIBONUCLEOSIDE, POTASSIUM ION, putative sugar kinase | | Authors: | Zhang, Y, Dougherty, M, Downs, D.M, Ealick, S.E. | | Deposit date: | 2004-07-09 | | Release date: | 2004-10-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of an Aminoimidazole Riboside Kinase from Salmonella enterica; Implications for the Evolution of the Ribokinase Superfamily
STRUCTURE, 12, 2004
|
|
1TYY
 
 | | Crystal structure of aminoimidazole riboside kinase from Salmonella enterica | | Descriptor: | POTASSIUM ION, putative sugar kinase | | Authors: | Zhang, Y, Dougherty, M, Downs, D.M, Ealick, S.E. | | Deposit date: | 2004-07-08 | | Release date: | 2004-10-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of an Aminoimidazole Riboside Kinase from Salmonella enterica; Implications for the Evolution of the Ribokinase Superfamily
STRUCTURE, 12, 2004
|
|
1TZ6
 
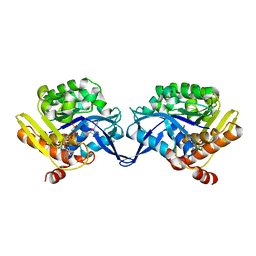 | | Crystal structure of aminoimidazole riboside kinase from Salmonella enterica complexed with aminoimidazole riboside and ATP analog | | Descriptor: | 5-AMINOIMIDAZOLE RIBONUCLEOSIDE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Zhang, Y, Dougherty, M, Downs, D.M, Ealick, S.E. | | Deposit date: | 2004-07-09 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of an Aminoimidazole Riboside Kinase from Salmonella enterica; Implications for the Evolution of the Ribokinase Superfamily
STRUCTURE, 12, 2004
|
|
3J32
 
 | | An asymmetric unit map from electron cryo-microscopy of Haliotis diversicolor molluscan hemocyanin isoform 1 (HdH1) | | Descriptor: | Hemocyanin isoform 1 | | Authors: | Zhang, Q, Dai, X, Cong, Y, Zhang, J, Chen, D.-H, Dougherty, M, Wang, J, Ludtke, S, Schmid, M.F, Chiu, W. | | Deposit date: | 2013-02-20 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of a molluscan hemocyanin suggests its allosteric mechanism.
Structure, 21, 2013
|
|
3IXW
 
 | | Scorpion Hemocyanin activated state pseudo atomic model built based on cryo-EM density map | | Descriptor: | Hemocyanin AA6 chain | | Authors: | Cong, Y, Zhang, Q, Woolford, D, Schweikardt, T, Khant, H, Ludtke, S, Chiu, W, Decker, H. | | Deposit date: | 2009-02-13 | | Release date: | 2009-06-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structural Mechanism of SDS-Induced Enzyme Activity of Scorpion Hemocyanin Revealed by Electron Cryomicroscopy.
Structure, 17, 2009
|
|
3IXV
 
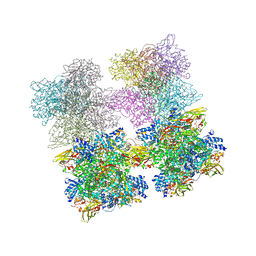 | | Scorpion Hemocyanin resting state pseudo atomic model built based on cryo-EM density map | | Descriptor: | Hemocyanin AA6 chain | | Authors: | Cong, Y, Zhang, Q, Woolford, D, Schweikardt, T, Khant, H, Ludtke, S, Chiu, W, Decker, H. | | Deposit date: | 2009-02-13 | | Release date: | 2009-06-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Structural Mechanism of SDS-Induced Enzyme Activity of Scorpion Hemocyanin Revealed by Electron Cryomicroscopy.
Structure, 17, 2009
|
|
3J03
 
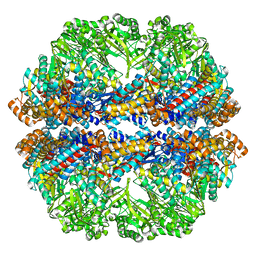 | | Lidless Mm-cpn in the closed state with ATP/AlFx | | Descriptor: | Lidless Mm-cpn | | Authors: | Zhang, J, Ma, B, DiMaio, F, Douglas, N.R, Joachimiak, L, Baker, D, Frydman, J, Levitt, M, Chiu, W. | | Deposit date: | 2011-02-10 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM structure of a group II chaperonin in the prehydrolysis ATP-bound state leading to lid closure.
Structure, 19, 2011
|
|
