4UY3
 
 | | Cytoplasmic domain of bacterial cell division protein ezra | | Descriptor: | SEPTATION RING FORMATION REGULATOR EZRA | | Authors: | Cleverley, R.M, Barrett, J.R, Basle, A, Khai-Bui, N, Hewitt, L, Solovyova, A, Xu, Z, Daniela, R.A, Dixon, N.E, Harry, E.J, Oakley, A.J, Vollmer, W, Lewis, R.J. | | Deposit date: | 2014-08-28 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Function of a Spectrin-Like Regulator of Bacterial Cytokinesis.
Nat.Commun., 5, 2014
|
|
2I05
 
 | | Escherichia Coli Replication Terminator Protein (Tus) Complexed With TerA DNA | | Descriptor: | 5'-D(*T*TP*AP*GP*TP*TP*AP*CP*AP*AP*CP*AP*TP*AP*CP*T)-3', 5'-D(*TP*AP*GP*TP*AP*TP*GP*TP*TP*GP*TP*AP*AP*CP*TP*A)-3', DNA replication terminus site-binding protein, ... | | Authors: | Oakley, A.J, Mulcair, M.D, Schaeffer, P.M, Dixon, N.E. | | Deposit date: | 2006-08-10 | | Release date: | 2007-08-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Polarity of Termination of DNA Replication in E. coli
To be Published
|
|
4UXV
 
 | | Cytoplasmic domain of bacterial cell division protein EzrA | | Descriptor: | SEPTATION RING FORMATION REGULATOR EZRA | | Authors: | Cleverley, R.M, Barrett, J.R, Basle, A, Khai-Bui, N, Hewitt, L, Solovyova, A, Xu, Z, Daniela, R.A, Dixon, N.E, Harry, E.J, Oakley, A.J, Vollmer, W, Lewis, R.J. | | Deposit date: | 2014-08-27 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.961 Å) | | Cite: | Structure and Function of a Spectrin-Like Regulator of Bacterial Cytokinesis.
Nat.Commun., 5, 2014
|
|
1QOR
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI QUINONE OXIDOREDUCTASE COMPLEXED WITH NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, QUINONE OXIDOREDUCTASE, SULFATE ION | | Authors: | Thorn, J.M, Barton, J.D, Dixon, N.E, Ollis, D.L, Edwards, K.J. | | Deposit date: | 1995-02-14 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Escherichia coli QOR quinone oxidoreductase complexed with NADPH.
J.Mol.Biol., 249, 1995
|
|
1T3W
 
 | | Crystal Structure of the E.coli DnaG C-terminal domain (residues 434 to 581) | | Descriptor: | ACETIC ACID, DNA primase | | Authors: | Oakley, A.J, Loscha, K.V, Schaeffer, P.M, Liepinsh, E, Wilce, M.C.J, Otting, G, Dixon, N.E. | | Deposit date: | 2004-04-28 | | Release date: | 2004-11-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal and solution structures of the helicase-binding domain of Escherichia coli primase
J.Biol.Chem., 280, 2005
|
|
2K7R
 
 | | N-terminal domain of the Bacillus subtilis helicase-loading protein DnaI | | Descriptor: | Primosomal protein dnaI, ZINC ION | | Authors: | Loscha, K.V, Jaudzems, K, Ioannou, C, Su, X.C, Hill, F.R, Otting, G, Dixon, N.E, Liepinsh, E. | | Deposit date: | 2008-08-19 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A novel zinc-binding fold in the helicase interaction domain of the Bacillus subtilis DnaI helicase loader
Nucleic Acids Res., 37, 2009
|
|
2I06
 
 | | Escherichia Coli Replication Terminator Protein (Tus) Complexed With DNA- Locked form | | Descriptor: | 5'-D(*T*TP*AP*GP*TP*TP*AP*CP*AP*AP*CP*AP*TP*AP*CP*T)-3', 5'-D(*TP*G*AP*TP*AP*TP*GP*TP*TP*GP*TP*AP*AP*CP*TP*A)-3', DNA replication terminus site-binding protein, ... | | Authors: | Oakley, A.J, Mulcair, M.D, Schaeffer, P.M, Dixon, N.E. | | Deposit date: | 2006-08-10 | | Release date: | 2007-08-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Polarity of Termination of DNA Replication in E. coli.
To be Published
|
|
4GX9
 
 | | Crystal structure of a DNA polymerase III alpha-epsilon chimera | | Descriptor: | DNA polymerase III subunit epsilon,DNA polymerase III subunit alpha | | Authors: | Li, N, Horan, N, Xu, Z.-Q, Jacques, D, Dixon, N.E, Oakley, A.J. | | Deposit date: | 2012-09-04 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Proofreading exonuclease on a tether: the complex between the E. coli DNA polymerase III subunits alpha, {varepsilon}, theta and beta reveals a highly flexible arrangement of the proofreading domain
Nucleic Acids Res., 41, 2013
|
|
3QSB
 
 | | Structure of E. coli polIIIbeta with (Z)-5-(1-((4'-Fluorobiphenyl-4-yl)methoxyimino)butyl)-2,2-dimethyl-4,6-dioxocyclohexanecarbonitrile | | Descriptor: | (1R,5R)-5-{(1Z)-N-[(4'-fluorobiphenyl-4-yl)methoxy]butanimidoyl}-2,2-dimethyl-4,6-dioxocyclohexanecarbonitrile, DNA polymerase III subunit beta | | Authors: | Wijffels, G, Johnson, W.M, Oakley, A.J, Turner, K, Epa, V.C, Briscoe, S.J, Polley, M, Liepa, A.J, Hofmann, A, Buchardt, J, Christensen, C, Prosselkov, P, Dalrymple, B.P, Alewood, P.F, Jennings, P.A, Dixon, N.E, Winkler, D.A. | | Deposit date: | 2011-02-20 | | Release date: | 2011-06-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding inhibitors of the bacterial sliding clamp by design
J.Med.Chem., 54, 2011
|
|
8GJ3
 
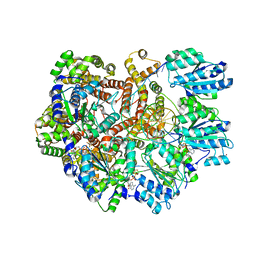 | | E. coli clamp loader on primed template DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA polymerase III subunit delta, DNA polymerase III subunit delta', ... | | Authors: | Oakley, A.J, Xu, Z.-Q, Dixon, N.E. | | Deposit date: | 2023-03-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural characterisation of the complete cycle of sliding clamp loading in Escherichia coli.
Nat Commun, 15, 2024
|
|
8GJ1
 
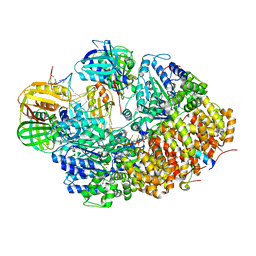 | | E. coli clamp loader with open clamp on primed template DNA (form 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Beta sliding clamp, DNA polymerase III subunit delta, ... | | Authors: | Oakley, A.J, Xu, Z.-Q, Dixon, N.E. | | Deposit date: | 2023-03-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural characterisation of the complete cycle of sliding clamp loading in Escherichia coli.
Nat Commun, 15, 2024
|
|
8GIZ
 
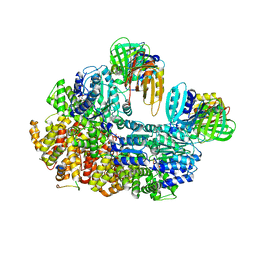 | | E. coli clamp loader with open clamp | | Descriptor: | Beta sliding clamp, DNA polymerase III subunit delta, DNA polymerase III subunit delta', ... | | Authors: | Oakley, A.J, Xu, Z.-Q, Dixon, N.E. | | Deposit date: | 2023-03-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural characterisation of the complete cycle of sliding clamp loading in Escherichia coli.
Nat Commun, 15, 2024
|
|
8GIY
 
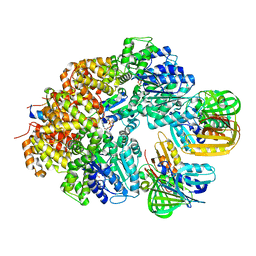 | | E. coli clamp loader with closed clamp | | Descriptor: | Beta sliding clamp, DNA polymerase III subunit delta, DNA polymerase III subunit delta', ... | | Authors: | Oakley, A.J, Xu, Z.-Q, Dixon, N.E. | | Deposit date: | 2023-03-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural characterisation of the complete cycle of sliding clamp loading in Escherichia coli.
Nat Commun, 15, 2024
|
|
8GJ0
 
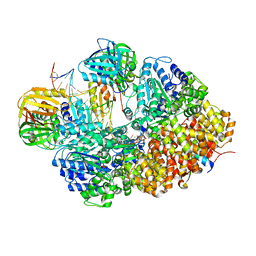 | | E. coli clamp loader with open clamp on primed template DNA (form 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Beta sliding clamp, DNA polymerase III subunit delta, ... | | Authors: | Oakley, A.J, Xu, Z.-Q, Dixon, N.E. | | Deposit date: | 2023-03-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural characterisation of the complete cycle of sliding clamp loading in Escherichia coli.
Nat Commun, 15, 2024
|
|
8GJ2
 
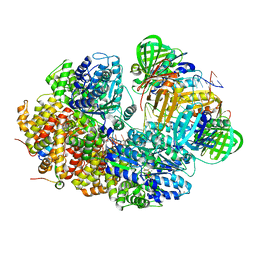 | | E. coli clamp loader with closed clamp on primed template DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Beta sliding clamp, DNA polymerase III subunit delta, ... | | Authors: | Oakley, A.J, Xu, Z.-Q, Dixon, N.E. | | Deposit date: | 2023-03-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural characterisation of the complete cycle of sliding clamp loading in Escherichia coli.
Nat Commun, 15, 2024
|
|
4GX8
 
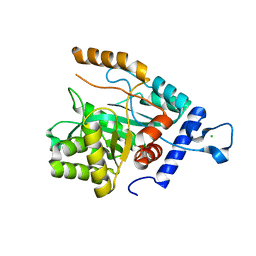 | | Crystal structure of a DNA polymerase III alpha-epsilon chimera | | Descriptor: | CHLORIDE ION, DNA polymerase III subunit epsilon,DNA polymerase III subunit alpha | | Authors: | Robinson, A, Horan, N, Xu, Z.-Q, Dixon, N.E, Oakley, A.J. | | Deposit date: | 2012-09-04 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Proofreading exonuclease on a tether: the complex between the E. coli DNA polymerase III subunits alpha, {varepsilon}, theta and beta reveals a highly flexible arrangement of the proofreading domain
Nucleic Acids Res., 41, 2013
|
|
1A16
 
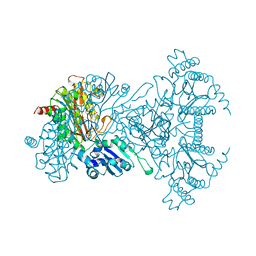 | | AMINOPEPTIDASE P FROM E. COLI WITH THE INHIBITOR PRO-LEU | | Descriptor: | AMINOPEPTIDASE P, LEUCINE, MANGANESE (II) ION, ... | | Authors: | Wilce, M.C, Bond, C.S, Lilley, P.E, Dixon, N.E, Freeman, H.C, Guss, J.M. | | Deposit date: | 1997-12-22 | | Release date: | 1999-04-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of a proline-specific aminopeptidase from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1TUH
 
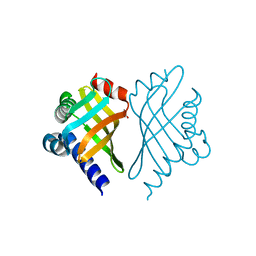 | | Structure of Bal32a from a Soil-Derived Mobile Gene Cassette | | Descriptor: | ACETATE ION, hypothetical protein EGC068 | | Authors: | Robinson, A, Wu, P.S.-C, Harrop, S.J, Schaeffer, P.M, Dixon, N.E, Gillings, M.R, Holmes, A.J, Nevalainen, K.M.H, Otting, G, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C. | | Deposit date: | 2004-06-25 | | Release date: | 2004-07-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Integron-associated Mobile Gene Cassettes Code for Folded Proteins: The Structure of Bal32a, a New Member of the Adaptable alpha+beta Barrel Family
J.Mol.Biol., 346, 2005
|
|
1MMI
 
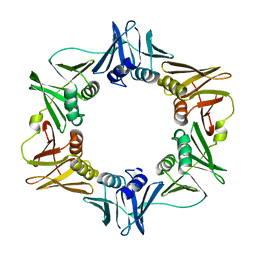 | | E. COLI DNA POLYMERASE BETA SUBUNIT | | Descriptor: | DNA polymerase III, beta chain | | Authors: | Oakley, A.J, Prosselkov, P, Wijffels, G, Beck, J.L, Wilce, M.C.J, Dixon, N.E. | | Deposit date: | 2002-09-04 | | Release date: | 2003-09-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Flexibility revealed by the 1.85 A crystal structure of the beta sliding-clamp subunit of Escherichia coli DNA polymerase III.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1DU2
 
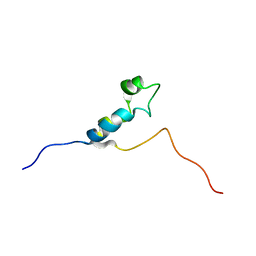 | | SOLUTION STRUCTURE OF THE THETA SUBUNIT OF DNA POLYMERASE III | | Descriptor: | DNA POLYMERASE III | | Authors: | Keniry, M.A, Berthon, H.A, Yang, J.-Y, Miles, C.S, Dixon, N.E. | | Deposit date: | 2000-01-13 | | Release date: | 2000-05-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the theta subunit of DNA polymerase III from Escherichia coli.
Protein Sci., 9, 2000
|
|
2EWJ
 
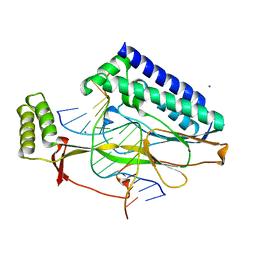 | | Escherichia Coli Replication Terminator Protein (Tus) Complexed With DNA- Locked form | | Descriptor: | 5'-D(*T*TP*AP*GP*TP*TP*AP*CP*AP*AP*CP*AP*TP*AP*CP*T)-3', 5'-D(*TP*G*AP*TP*AP*TP*GP*TP*TP*GP*TP*AP*AP*CP*TP*A)-3', DNA replication terminus site-binding protein, ... | | Authors: | Oakley, A.J, Mulcair, M.D, Schaeffer, P.M, Dixon, N.E. | | Deposit date: | 2005-11-03 | | Release date: | 2006-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A molecular mousetrap determines polarity of termination of DNA replication in E. coli.
Cell(Cambridge,Mass.), 125, 2006
|
|
2GUI
 
 | | Structure and Function of Cyclized Versions of the Proofreading Exonuclease Subunit of E. coli DNA Polymerase III | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA polymerase III epsilon subunit, ... | | Authors: | Park, A.Y, Carr, P.D, Ollis, D.L, Dixon, N.E. | | Deposit date: | 2006-04-30 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Function of Cyclized Versions of the Proofreading Exonuclease Subunit E. coli DNA Polymerase III
To be Published
|
|
2HAJ
 
 | |
1J53
 
 | | Structure of the N-terminal Exonuclease Domain of the Epsilon Subunit of E.coli DNA Polymerase III at pH 8.5 | | Descriptor: | 1,2-ETHANEDIOL, DNA polymerase III, epsilon chain, ... | | Authors: | Hamdan, S, Carr, P.D, Brown, S.E, Ollis, D.L, Dixon, N.E. | | Deposit date: | 2002-01-22 | | Release date: | 2002-10-16 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Proofreading during Replication of the Escherichia coli Chromosome
Structure, 10, 2002
|
|
1J54
 
 | | Structure of the N-terminal exonuclease domain of the epsilon subunit of E.coli DNA polymerase III at pH 5.8 | | Descriptor: | 1,2-ETHANEDIOL, DNA polymerase III, epsilon chain, ... | | Authors: | Hamdan, S, Carr, P.D, Brown, S.E, Ollis, D.L, Dixon, N.E. | | Deposit date: | 2002-01-22 | | Release date: | 2002-10-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Proofreading during Replication of the Escherichia coli Chromosome
Structure, 10, 2002
|
|
