1BA7
 
 | | SOYBEAN TRYPSIN INHIBITOR | | Descriptor: | TRYPSIN INHIBITOR (KUNITZ) | | Authors: | De Meester, P, Brick, P, Lloyd, L.F, Blow, D.M, Onesti, S. | | Deposit date: | 1998-04-22 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Kunitz-type soybean trypsin inhibitor (STI): implication for the interactions between members of the STI family and tissue-plasminogen activator.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1A62
 
 | | CRYSTAL STRUCTURE OF THE RNA-BINDING DOMAIN OF THE TRANSCRIPTIONAL TERMINATOR PROTEIN RHO | | Descriptor: | RHO | | Authors: | Allison, T.J, Wood, T.C, Briercheck, D.M, Rastinejad, F, Richardson, J.P, Rule, G.S. | | Deposit date: | 1998-03-05 | | Release date: | 1998-06-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the RNA-binding domain from transcription termination factor rho.
Nat.Struct.Biol., 5, 1998
|
|
1A63
 
 | | THE NMR STRUCTURE OF THE RNA BINDING DOMAIN OF E.COLI RHO FACTOR SUGGESTS POSSIBLE RNA-PROTEIN INTERACTIONS, 10 STRUCTURES | | Descriptor: | RHO | | Authors: | Briercheck, D.M, Wood, T.C, Allison, T.J, Richardson, J.P, Rule, G.S. | | Deposit date: | 1998-03-05 | | Release date: | 1998-05-27 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the RNA binding domain of E. coli rho factor suggests possible RNA-protein interactions.
Nat.Struct.Biol., 5, 1998
|
|
6Q13
 
 | | CRYSTAL STRUCTURE OF LDHA IN COMPLEX WITH COMPOUND NCGC00420737-09 AT 2.00 A RESOLUTION | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-[5-(cyclopropylmethyl)-4-[(3-fluoro-4-sulfamoylphenyl)methyl]-3-{3-[(5-methylthiophen-2-yl)ethynyl]phenyl}-1H-pyrazol-1-yl]-1,3-thiazole-4-carboxylic acid, ... | | Authors: | Davies, D.R, Dranow, D.M. | | Deposit date: | 2019-08-02 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pyrazole-Based Lactate Dehydrogenase Inhibitors with Optimized Cell Activity and Pharmacokinetic Properties.
J.Med.Chem., 63, 2020
|
|
1B4C
 
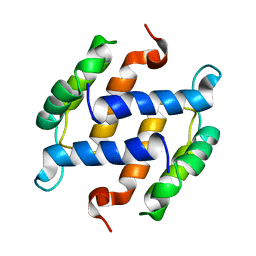 | | SOLUTION STRUCTURE OF RAT APO-S100B USING DIPOLAR COUPLINGS | | Descriptor: | PROTEIN (S-100 PROTEIN, BETA CHAIN) | | Authors: | Weber, D.J, Drohat, A.C, Tjandra, N, Baldisseri, D.M. | | Deposit date: | 1998-12-17 | | Release date: | 1998-12-30 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The use of dipolar couplings for determining the solution structure of rat apo-S100B(betabeta).
Protein Sci., 8, 1999
|
|
1BPS
 
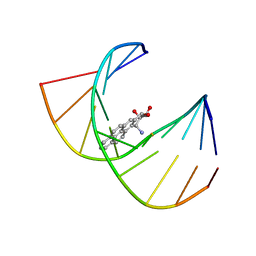 | | MINOR CONFORMER OF A BENZO[A]PYRENE DIOL EPOXIDE ADDUCT OF DA IN DUPLEX DNA | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA (5'-D(*CP*TP*CP*GP*GP*GP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*CP*(BAP)AP*CP*GP*AP*G)-3') | | Authors: | Schwartz, J.S, Rice, J.S, Luxon, B.A, Sayer, J.M, Xie, G, Yeh, H.J.C, Liu, X, Jerina, D.M, Gorenstein, D.G. | | Deposit date: | 1998-08-06 | | Release date: | 1998-08-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the minor conformer of a DNA duplex containing a dG mismatch opposite a benzo[a]pyrene diol epoxide/dA adduct: glycosidic rotation from syn to anti at the modified deoxyadenosine.
Biochemistry, 36, 1997
|
|
1BQ8
 
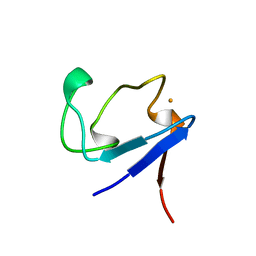 | | Rubredoxin (Methionine Mutant) from Pyrococcus Furiosus | | Descriptor: | FE (III) ION, PROTEIN (RUBREDOXIN) | | Authors: | Bau, R, Rees, D.C, Kurtz, D.M, Scott, R.A, Huang, H, Adams, M.W.W, Eidsness, M.K. | | Deposit date: | 1998-08-22 | | Release date: | 1998-08-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal Structure of Rubredoxin from Pyrococcus Furiosus at 0.95 Angstroms Resolution, and the structures of N-terminal methionine and formylmethionine variants of Pf Rd. Contributions of N-terminal interactions to thermostability
J.BIOL.INORG.CHEM., 3, 1998
|
|
1BRF
 
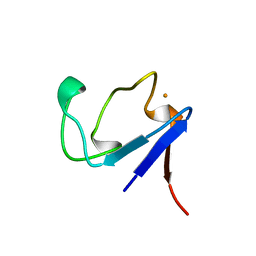 | | Rubredoxin (Wild Type) from Pyrococcus Furiosus | | Descriptor: | FE (III) ION, PROTEIN (RUBREDOXIN) | | Authors: | Bau, R, Rees, D.C, Kurtz, D.M, Scott, R.A, Huang, H, Adams, M.W.W, Eidsness, M.K. | | Deposit date: | 1998-08-24 | | Release date: | 1998-09-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Crystal Structure of Rubredoxin from Pyrococcus Furiosus at 0.95 Angstroms Resolution, and the structures of N-terminal methionine and formylmethionine variants of Pf Rd. Contributions of N-terminal interactions to thermostability
J.BIOL.INORG.CHEM., 3, 1998
|
|
1BQ9
 
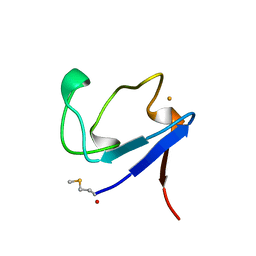 | | Rubredoxin (Formyl Methionine Mutant) from Pyrococcus Furiosus | | Descriptor: | FE (III) ION, PROTEIN (RUBREDOXIN) | | Authors: | Bau, R, Rees, D.C, Kurtz, D.M, Scott, R.A, Huang, H, Adams, M.W.W, Eidsness, M.K. | | Deposit date: | 1998-08-22 | | Release date: | 1998-08-26 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure of Rubredoxin from Pyrococcus Furiosus at 0.95 Angstroms Resolution, and the structures of N-terminal methionine and formylmethionine variants of Pf Rd. Contributions of N-terminal interactions to thermostability
J.BIOL.INORG.CHEM., 3, 1998
|
|
8U7Z
 
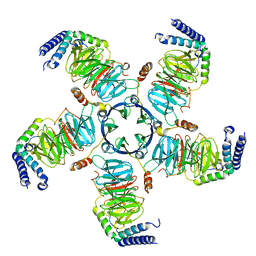 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: Local Refinment of KCTD5(CTD)/Gbeta1gamma2 | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/Cullin3/G beta gamma E3 ubiquitin ligase complex
Proc.Natl.Acad.Sci.USA, 2024
|
|
8U80
 
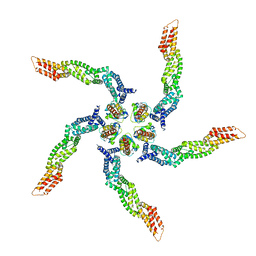 | |
8U83
 
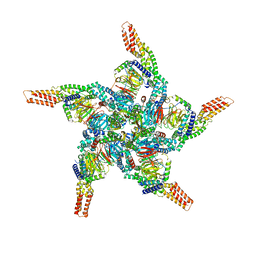 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State C From Composite RELION Multi-body Refinement Map | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.975 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/Cullin3/G beta gamma E3 ubiquitin ligase complex
Proc.Natl.Acad.Sci.USA, 2024
|
|
8U84
 
 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State D From Composite RELION Multi-body Refinement Map | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/Cullin3/G beta gamma E3 ubiquitin ligase complex
Proc.Natl.Acad.Sci.USA, 2024
|
|
8U82
 
 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State B From Composite RELION Multi-body Refinement Map | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/Cullin3/G beta gamma E3 ubiquitin ligase complex
Proc.Natl.Acad.Sci.USA, 2024
|
|
8U81
 
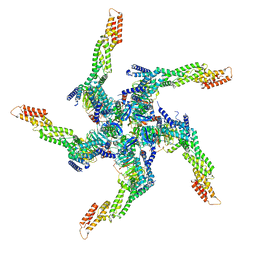 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State A From Composite RELION Multi-body Refinement Map | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/Cullin3/G beta gamma E3 ubiquitin ligase complex
Proc.Natl.Acad.Sci.USA, 2024
|
|
6XDC
 
 | |
1AFO
 
 | |
1ATG
 
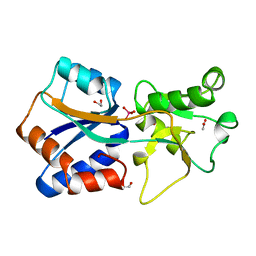 | | AZOTOBACTER VINELANDII PERIPLASMIC MOLYBDATE-BINDING PROTEIN | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, PERIPLASMIC MOLYBDATE-BINDING PROTEIN, ... | | Authors: | Lawson, D.M, Pau, R.N, Williams, C.E.M, Mitchenall, L.A. | | Deposit date: | 1997-08-14 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Ligand size is a major determinant of specificity in periplasmic oxyanion-binding proteins: the 1.2 A resolution crystal structure of Azotobacter vinelandii ModA.
Structure, 6, 1998
|
|
4TS1
 
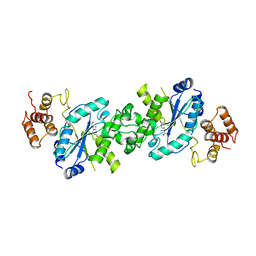 | |
8PFC
 
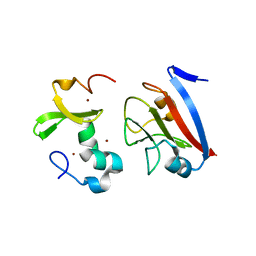 | | Crystal structure of binary complex between Aster yellows witches'-broom phytoplasma effector SAP05 and the zinc finger domain of SPL5 from Arabidopsis thaliana | | Descriptor: | Sequence-variable mosaic (SVM) signal sequence domain-containing protein, Squamosa promoter-binding-like protein 5, ZINC ION | | Authors: | Huang, W, Liu, Q, Maqbool, A, Stevenson, C.E.M, Lawson, D.M, Kamoun, S, Hogenhout, S.A. | | Deposit date: | 2023-06-15 | | Release date: | 2023-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bimodular architecture of bacterial effector SAP05 that drives ubiquitin-independent targeted protein degradation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8PFD
 
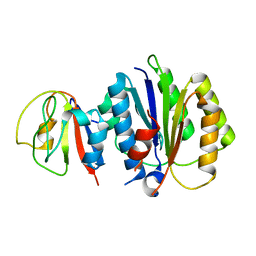 | | Crystal structure of binary complex between Aster yellows witches'-broom phytoplasma effector SAP05 and the von Willebrand Factor Type A domain of the proteasomal ubiquitin receptor Rpn10 from Arabidopsis thaliana | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4 homolog, Sequence-variable mosaic (SVM) signal sequence domain-containing protein | | Authors: | Huang, W, Liu, Q, Maqbool, A, Stevenson, C.E.M, Lawson, D.M, Kamoun, S, Hogenhout, S.A. | | Deposit date: | 2023-06-15 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Bimodular architecture of bacterial effector SAP05 that drives ubiquitin-independent targeted protein degradation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1BFY
 
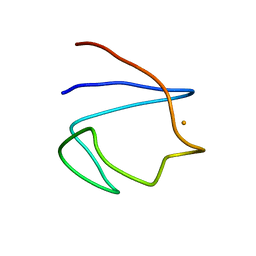 | | SOLUTION STRUCTURE OF REDUCED CLOSTRIDIUM PASTEURIANUM RUBREDOXIN, NMR, 20 STRUCTURES | | Descriptor: | FE (III) ION, RUBREDOXIN | | Authors: | Bertini, I, Kurtz Junior, D.M, Eidsness, M.K, Liu, G, Luchinat, C, Rosato, A, Scott, R.A. | | Deposit date: | 1998-05-23 | | Release date: | 1999-05-25 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Reduced Clostridium Pasteurianum Rubredoxin
J.Biol.Inorg.Chem., 3, 1998
|
|
1BUY
 
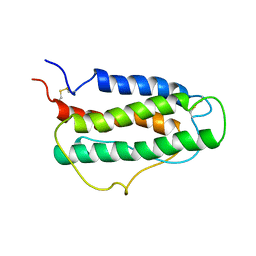 | | HUMAN ERYTHROPOIETIN, NMR MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PROTEIN (ERYTHROPOIETIN) | | Authors: | Cheetham, J.C, Smith, D.M, Aoki, K.H, Stevenson, J.L, Hoeffel, T.J, Syed, R.S, Egrie, J, Harvey, T.S. | | Deposit date: | 1998-09-08 | | Release date: | 1999-09-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of human erythropoietin and a comparison with its receptor bound conformation.
Nat.Struct.Biol., 5, 1998
|
|
1BVB
 
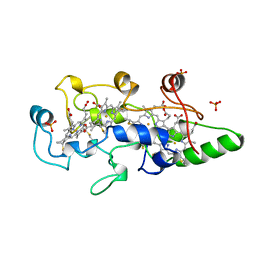 | | HEME-PACKING MOTIFS REVEALED BY THE CRYSTAL STRUCTURE OF CYTOCHROME C554 FROM NITROSOMONAS EUROPAEA | | Descriptor: | CYTOCHROME C-554, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Iverson, T.M, Arciero, D.M, Hsu, B.T, Logan, M.S.P, Hooper, A.B, Rees, D.C. | | Deposit date: | 1998-09-16 | | Release date: | 1999-05-18 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Heme packing motifs revealed by the crystal structure of the tetra-heme cytochrome c554 from Nitrosomonas europaea.
Nat.Struct.Biol., 5, 1998
|
|
1BJ2
 
 | |
