3DM8
 
 | | Crystal Structure of Putative Isomerase from Rhodopseudomonas palustris | | Descriptor: | DODECYL NONA ETHYLENE GLYCOL ETHER, uncharacterized protein RPA4348 | | Authors: | Cymborowski, M, Chruszcz, M, Skarina, T, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-06-30 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Putative Isomerase from Rhodopseudomonas palustris
To be Published
|
|
4KJM
 
 | | Crystal structure of the Staphylococcus aureus protein (NP_646141.1, domain 3912-4037) similar to streptococcal adhesins emb and ebhA/ebhB | | Descriptor: | ACETATE ION, CHLORIDE ION, Extracellular matrix-binding protein ebh, ... | | Authors: | Cymborowski, M, Shabalin, I.G, Joachimiak, G, Chruszcz, M, Gornicki, P, Zhang, R, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-03 | | Release date: | 2013-05-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the Staphylococcus aureus protein (NP_646141.1, domain 3912-4037) similar to streptococcal adhesins emb and ebhA/ebhB
To be Published
|
|
3I4P
 
 | | Crystal structure of AsnC family transcriptional regulator from Agrobacterium tumefaciens | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Cymborowski, M, Chruszcz, M, Luo, H, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-02 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of AsnC family transcriptional regulator from Agrobacterium tumefaciens
To be Published
|
|
2PD0
 
 | | Protein cgd2_2020 from Cryptosporidium parvum | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Hypothetical protein | | Authors: | Cymborowski, M, Chruszcz, M, Hills, T, Lew, J, Melone, M, Zhao, Y, Artz, J, Wernimont, A, Edwards, A, Sundstrom, M, Weigelt, J, Bochkarev, A, Arrowsmith, C, Hui, R, Minor, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-30 | | Release date: | 2007-04-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein cgd2_2020 from Cryptosporidium parvum
To be Published
|
|
2G3A
 
 | | Crystal structure of putative acetyltransferase from Agrobacterium tumefaciens | | Descriptor: | acetyltransferase | | Authors: | Cymborowski, M, Xu, X, Chruszcz, M, Zheng, H, Gu, J, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-17 | | Release date: | 2006-03-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of putative acetyltransferase from Agrobacterium tumefaciens
To be Published
|
|
2Q24
 
 | | Crystal structure of TetR transcriptional regulator SCO0520 from Streptomyces coelicolor | | Descriptor: | ACETATE ION, CHLORIDE ION, Putative tetR family transcriptional regulator | | Authors: | Cymborowski, M, Chruszcz, M, Koclega, K.D, Filippova, E.V, Xu, X, Gu, J, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-05-25 | | Release date: | 2007-07-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a putative transcriptional regulator SCO0520 from Streptomyces coelicolor A3(2) reveals an unusual dimer among TetR family proteins.
J.Struct.Funct.Genom., 12, 2011
|
|
3SLB
 
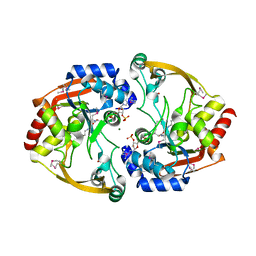 | | Crystal structure of BA2930 in complex with AcCoA and cytosine | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 6-AMINOPYRIMIDIN-2(1H)-ONE, ... | | Authors: | Klimecka, M.M, Chruszcz, M, Porebski, P.J, Cymborowski, M, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-06-24 | | Release date: | 2011-07-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of BA2930 in complex with AcCoA and cytosine
TO BE PUBLISHED
|
|
3SLF
 
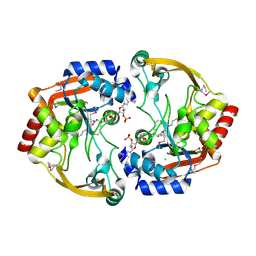 | | Crystal structure of BA2930 in complex with AcCoA and uracil | | Descriptor: | ACETYL COENZYME *A, Aminoglycoside N3-acetyltransferase, CHLORIDE ION, ... | | Authors: | Klimecka, M.M, Chruszcz, M, Porebski, P.J, Cymborowski, M, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-06-24 | | Release date: | 2011-07-20 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of BA2930 in complex with AcCoA and uracil
TO BE PUBLISHED
|
|
4XOX
 
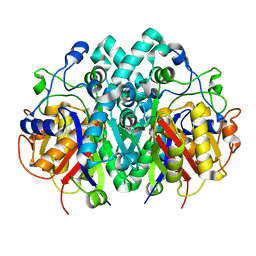 | | Structure of beta-ketoacyl-ACP synthase I (FabB) from Vibrio Cholerae | | Descriptor: | 3-oxoacyl-ACP synthase | | Authors: | Hou, J, Grabowski, M, Cymborowski, M, Zheng, H, Cooper, D.R, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-01-16 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of beta-ketoacyl-ACP synthase I (FabB) from Vibrio Cholerae
to be published
|
|
1Y89
 
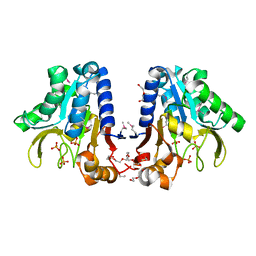 | | Crystal Structure of devB protein | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, DI(HYDROXYETHYL)ETHER, NONAETHYLENE GLYCOL, ... | | Authors: | Lazarski, K, Cymborowski, M, Chruszcz, M, Zheng, H, Zhang, R, Lezondra, L, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-10 | | Release date: | 2005-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of devB protein
To be Published
|
|
3E4F
 
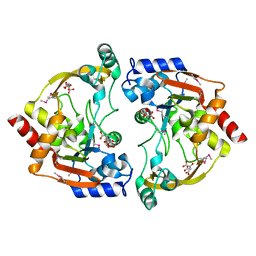 | | Crystal structure of BA2930- a putative aminoglycoside N3-acetyltransferase from Bacillus anthracis | | Descriptor: | Aminoglycoside N3-acetyltransferase, CITRIC ACID | | Authors: | Klimecka, M.M, Chruszcz, M, Skarina, T, Onopryienko, O, Cymborowski, M, Savchenko, A, Edwards, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-08-11 | | Release date: | 2008-08-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of a Putative Aminoglycoside N-Acetyltransferase from Bacillus anthracis.
J.Mol.Biol., 410, 2011
|
|
1XVI
 
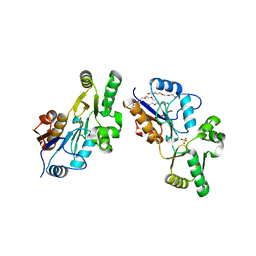 | | Crystal Structure of YedP, phosphatase-like domain protein from Escherichia coli K12 | | Descriptor: | PENTAETHYLENE GLYCOL, Putative mannosyl-3-phosphoglycerate phosphatase, SULFATE ION, ... | | Authors: | Kim, Y, Joachimiak, A, Cymborowski, M, Skarina, T, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-10-28 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal Structure of YedP, phosphatase-like domain protein from Escherichia coli K12
To be Published
|
|
3FZV
 
 | | Crystal structure of PA01 protein, putative LysR family transcriptional regulator from Pseudomonas aeruginosa | | Descriptor: | Probable transcriptional regulator, SULFATE ION | | Authors: | Knapik, A.A, Tkaczuk, K.L, Chruszcz, M, Wang, S, Zimmerman, M.D, Cymborowski, M, Skarina, T, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Bujnicki, J.M, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-26 | | Release date: | 2009-03-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structure of PA01 protein, putative LysR family transcriptional regulator from Pseudomonas aeruginosa
To be Published
|
|
3TL2
 
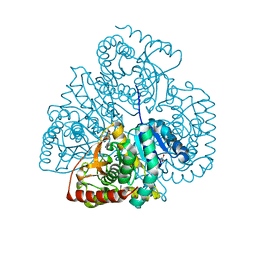 | | Crystal structure of Bacillus anthracis str. Ames malate dehydrogenase in closed conformation. | | Descriptor: | 1,2-ETHANEDIOL, Malate dehydrogenase, THIOCYANATE ION | | Authors: | Blus, B.J, Chruszcz, M, Tkaczuk, K.L, Osinski, T, Cymborowski, M, Kudritska, M, Grimshaw, S, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-08-29 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Bacillus anthracis str. Ames malate dehydrogenase in closed conformation.
TO BE PUBLISHED
|
|
4JWV
 
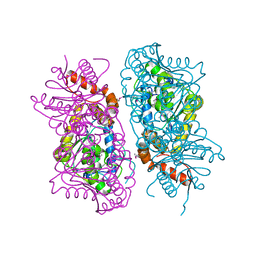 | | Crystal Structure of putative short chain enoyl-CoA hydratase from Novosphingobium aromaticivorans DSM 12444 | | Descriptor: | Short chain enoyl-CoA hydratase | | Authors: | Cooper, D.R, Mikolajczak, K, Cymborowski, M, Grabowski, M, Ahmed, M, Stead, M, Hillerich, B, Seidel, R, Zimmerman, M, Bonanno, J.B, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-27 | | Release date: | 2013-05-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of putative short chain enoyl-CoA hydratase from Novosphingobium aromaticivorans DSM 12444
To be Published
|
|
1SFX
 
 | | X-ray crystal structure of putative HTH transcription regulator from Archaeoglobus fulgidus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Conserved hypothetical protein AF2008 | | Authors: | Osipiuk, J, Skarina, T, Savchenko, A, Edwards, A, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-20 | | Release date: | 2004-08-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray crystal structure of putative HTH transcription regulator from Archaeoglobus fulgidus
To be Published
|
|
1T9K
 
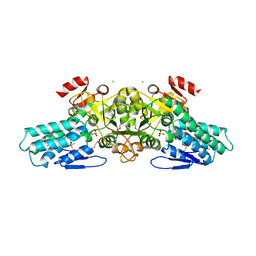 | | X-ray crystal structure of aIF-2B alpha subunit-related translation initiation factor [Thermotoga maritima] | | Descriptor: | CHLORIDE ION, Probable methylthioribose-1-phosphate isomerase, SULFATE ION | | Authors: | Osipiuk, J, Skarina, T, Savchenko, A, Edwards, A, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-17 | | Release date: | 2004-06-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray crystal structure of aIF-2B translation initiation factor from Thermotoga maritima
To be Published
|
|
1TU1
 
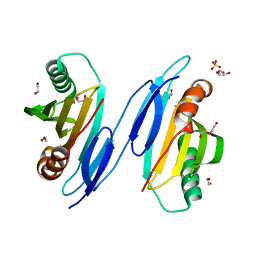 | | Crystal Structure of Protein of Unknown Function PA94 from Pseudomonas aeruginosa, Putative Regulator | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Osipiuk, J, Evdokimova, E, Savchenko, A, Edwards, A, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-06-24 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray crystal structure of hypothetical protein PA94 from Pseudomonas aeruginosa
To be Published
|
|
4JRR
 
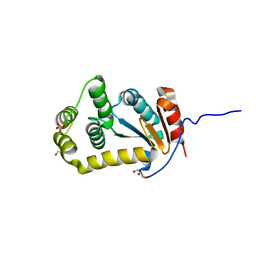 | | Crystal structure of disulfide bond oxidoreductase DsbA1 from Legionella pneumophila | | Descriptor: | GLYCEROL, SULFATE ION, Thiol:disulfide interchange protein DsbA | | Authors: | Shumilin, I.A, Jameson-Lee, M, Cymborowski, M, Domagalski, M.J, Chertihin, O, Kpadeh, Z.Z, Yeh, A.J, Hoffman, P.S, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-21 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of disulfide bond oxidoreductase DsbA1 from Legionella pneumophila
To be Published
|
|
2NP3
 
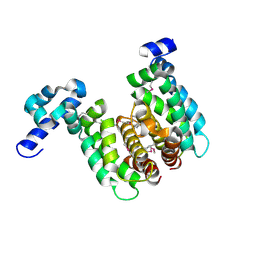 | | Crystal structure of TetR-family regulator (SCO0857) from Streptomyces coelicolor A3. | | Descriptor: | Putative TetR-family regulator | | Authors: | Koclega, K.D, Xu, X, Chruszcz, M, Gu, J, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-10-26 | | Release date: | 2006-11-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of TetR-family regulator (SCO0857) from Streptomyces coelicolor A3.
To be Published
|
|
2NP5
 
 | | Crystal structure of a transcriptional regulator (RHA1_ro04179) from Rhodococcus sp. Rha1. | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, Transcriptional regulator | | Authors: | Chruszcz, M, Evdokimova, E, Kagan, O, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-10-26 | | Release date: | 2006-11-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a transcriptional regulator (RHA1_ro04179) from Rhodococcus sp. Rha1.
TO BE PUBLISHED
|
|
7KYJ
 
 | | Structure of a GNAT superfamily PA3944 acetyltransferase in complex with zinc | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Acetyltransferase PA3944, ... | | Authors: | Czub, M.P, Porebski, P.J, Cymborowski, M, Shabalin, I.G, Reidl, C.T, Becker, D.P, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a GNAT superfamily PA3944 acetyltransferase in complex with zinc
To Be Published
|
|
7KYE
 
 | | Structure of a GNAT superfamily PA3944 acetyltransferase in complex with CHES | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Acetyltransferase PA3944, ... | | Authors: | Czub, M.P, Porebski, P.J, Cymborowski, M, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of a GNAT superfamily PA3944 acetyltransferase in complex with CHES
To Be Published
|
|
3IBZ
 
 | | Crystal structure of putative tellurium resistant like protein (TerD) from Streptomyces coelicolor A3(2) | | Descriptor: | CALCIUM ION, Putative tellurium resistant like protein TerD, SULFATE ION | | Authors: | Klimecka, M, Chruszcz, M, Cymborowski, M, Xu, X, Cui, H, Joachimiak, A, Edwards, A, Savchenko, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-17 | | Release date: | 2009-08-18 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of putative tellurium resistant like protein (TerD) from Streptomyces coelicolor A3(2)
To be Published
|
|
3KXR
 
 | | Structure of the cystathionine beta-synthase pair domain of the putative Mg2+ transporter SO5017 from Shewanella oneidensis MR-1. | | Descriptor: | CHLORIDE ION, Magnesium transporter, putative | | Authors: | Fratczak, Z, Zimmerman, M.D, Chruszcz, M, Cymborowski, M, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-12-03 | | Release date: | 2009-12-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure of the cystathionine beta-synthase pair domain of the putative Mg2+ transporter SO5017 from Shewanella oneidensis MR-1.
To be Published
|
|
