5A7V
 
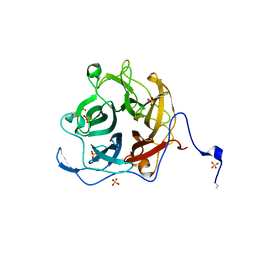 | | The GH130 family of mannoside phosphorylases contains glycoside hydrolases that target beta-1,2 mannosidic linkages in Candida mannan | | Descriptor: | PUTATIVE GLYCOSIDASE PH117-RELATED, SULFATE ION, alpha-D-mannopyranose, ... | | Authors: | Cuskin, F, Basle, A, Day, A.M, Ladeveze, S, Potocki-Veronese, G, Davies, G.J, Gilbert, H.J, Lowe, E. | | Deposit date: | 2015-07-10 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Gh130 Family of Mannoside Phosphorylases Contains Glycoside Hydrolases that Target Beta-1,2 Mannosidic Linkages in Candida Mannan
J.Biol.Chem., 290, 2015
|
|
4UTF
 
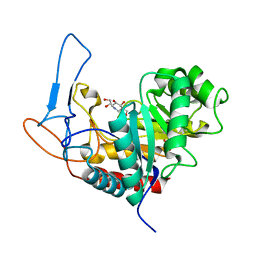 | | Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-isofagomine and alpha- 1,2-mannobiose | | Descriptor: | 1,2-ETHANEDIOL, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, GLYCOSYL HYDROLASE FAMILY 71, ... | | Authors: | Cuskin, F, Lowe, E.C, Temple, M.J, Zhu, Y, Pudlo, N.A, Cameron, E.A, Urs, K, Thompson, A.J, Cartmell, A, Rogowski, A, Tolbert, T, Piens, K, Bracke, D, Vervecken, W, Hakki, Z, Speciale, G, Munoz-Munoz, J.L, Pena, M.J, McLean, R, Suits, M.D, Boraston, A.B, Atherly, T, Ziemer, C.J, Williams, S.J, Davies, G.J, Abbott, D.W, Martens, E.C, Gilbert, H.J. | | Deposit date: | 2014-07-21 | | Release date: | 2014-12-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Human Gut Bacteroidetes Can Utilize Yeast Mannan Through a Selfish Mechanism.
Nature, 517, 2015
|
|
4B1L
 
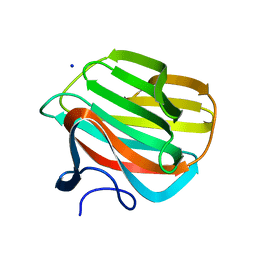 | | CARBOHYDRATE BINDING MODULE CBM66 FROM BACILLUS SUBTILIS | | Descriptor: | LEVANASE, SODIUM ION, beta-D-fructofuranose | | Authors: | Cuskin, F, Flint, J.E, Morland, C, Basle, A, Henrissat, B, Countinho, P.M, Strazzulli, A, Solzehinkin, A, Davies, G.J, Gilbert, H.J, Gloster, T.M. | | Deposit date: | 2012-07-11 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | How Nature Can Exploit Nonspecific Catalytic and Carbohydrate Binding Modules to Create Enzymatic Specificity
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4B1M
 
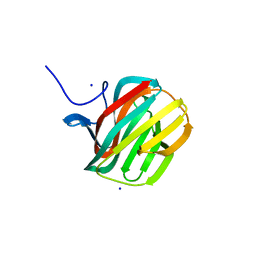 | | CARBOHYDRATE BINDING MODULE CBM66 FROM BACILLUS SUBTILIS | | Descriptor: | LEVANASE, SODIUM ION, SULFATE ION, ... | | Authors: | Cuskin, F, Flint, J.E, Morland, C, Basle, A, Henrissat, B, Countinho, P.M, Strazzulli, A, Solzehinkin, A, Davies, G.J, Gilbert, H.J, Gloster, T.M. | | Deposit date: | 2012-07-11 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | How Nature Can Exploit Nonspecific Catalytic and Carbohydrate Binding Modules to Create Enzymatic Specificity
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4C1S
 
 | | Glycoside hydrolase family 76 (mannosidase) Bt3792 from Bacteroides thetaiotaomicron VPI-5482 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, GLYCOSIDE HYDROLASE FAMILY 76 MANNOSIDASE | | Authors: | Cuskin, F, Lowe, E.C, Zhu, Y, Temple, M, Thompson, A.J, Cartmell, A, Piens, K, Bracke, D, Vervecken, W, Munoz-Munoz, J.L, Suits, M.D.L, Boraston, A.B, Williams, S.J, Davies, G.J, Abbott, W.D, Martens, E.C, Gilbert, H.J. | | Deposit date: | 2013-08-13 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human Gut Bacteroidetes Can Utilize Yeast Mannan Through a Selfish Mechanism.
Nature, 517, 2015
|
|
4C1R
 
 | | Bacteroides thetaiotaomicron VPI-5482 mannosyl-6-phosphatase Bt3783 | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, MANNOSYL-6-PHOSPHATASE | | Authors: | Cuskin, F, Lowe, E.C, Zhu, Y, Temple, M, Thompson, A.J, Cartmell, A, Piens, K, Bracke, D, Vervecken, W, Munoz-Munoz, J.L, Suits, M.D.L, Boraston, A.B, Williams, S.J, Davies, G.J, Abbott, W.D, Martens, E.C, Gilbert, H.J. | | Deposit date: | 2013-08-13 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human Gut Bacteroidetes Can Utilize Yeast Mannan Through a Selfish Mechanism.
Nature, 517, 2015
|
|
4V1S
 
 | | Structure of the GH76 alpha-mannanase BT2949 from Bacteroides thetaiotaomicron | | Descriptor: | ALPHA-1,6-MANNANASE, GLYCEROL | | Authors: | Thompson, A.J, Cuskin, F, Spears, R.J, Dabin, J, Turkenburg, J.P, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2014-10-02 | | Release date: | 2015-02-11 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Gh76 Alpha-Mannanase Homolog, Bt2949, from the Gut Symbiont Bacteroides Thetaiotaomicron
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4V1R
 
 | | Structure of a selenomethionine derivative of the GH76 alpha- mannanase BT2949 Bacteroides thetaiotaomicron | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-1,6-MANNANASE, S,R MESO-TARTARIC ACID | | Authors: | Thompson, A.J, Cuskin, F, Spears, R.J, Dabin, J, Turkenburg, J.P, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2014-10-02 | | Release date: | 2015-02-11 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Gh76 Alpha-Mannanase Homolog, Bt2949, from the Gut Symbiont Bacteroides Thetaiotaomicron
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5NGK
 
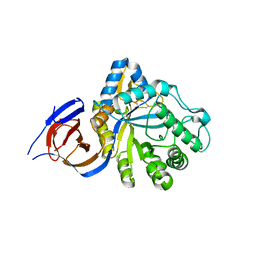 | | The endo-beta1,6-glucanase BT3312 | | Descriptor: | Glucosylceramidase | | Authors: | Basle, A, Temple, M, Cuskin, F, Lowe, E, Gilbert, H. | | Deposit date: | 2017-03-17 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Bacteroidetes locus dedicated to fungal 1,6-beta-glucan degradation: Unique substrate conformation drives specificity of the key endo-1,6-beta-glucanase.
J. Biol. Chem., 292, 2017
|
|
5LA1
 
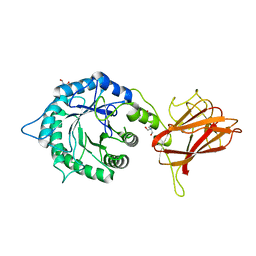 | | The mechanism by which arabinoxylanases can recognise highly decorated xylans | | Descriptor: | CALCIUM ION, Carbohydrate binding family 6, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM, ... | | Authors: | Basle, A, Labourel, A, Cuskin, F, Jackson, A, Crouch, L, Rogowski, A, Gilbert, H. | | Deposit date: | 2016-06-13 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Mechanism by Which Arabinoxylanases Can Recognize Highly Decorated Xylans.
J.Biol.Chem., 291, 2016
|
|
5LA2
 
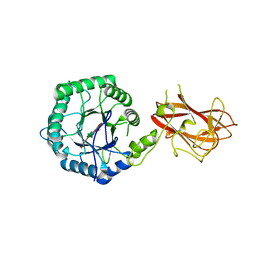 | | The mechanism by which arabinoxylanases can recognise highly decorated xylans | | Descriptor: | CALCIUM ION, Carbohydrate binding family 6, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-[alpha-L-arabinofuranose-(1-3)]alpha-D-xylopyranose, ... | | Authors: | Basle, A, Labourel, A, Cuskin, F, Jackson, A, Crouch, L, Rogowski, A, Gilbert, H. | | Deposit date: | 2016-06-13 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Mechanism by Which Arabinoxylanases Can Recognize Highly Decorated Xylans.
J.Biol.Chem., 291, 2016
|
|
5LA0
 
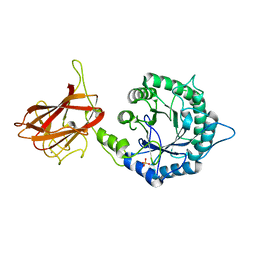 | | The mechanism by which arabinoxylanases can recognise highly decorated xylans | | Descriptor: | CALCIUM ION, Carbohydrate binding family 6, SULFATE ION, ... | | Authors: | Basle, A, Labourel, A, Cuskin, F, Jackson, A, Crouch, L, Rogowski, A, Gilbert, A. | | Deposit date: | 2016-06-13 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Mechanism by Which Arabinoxylanases Can Recognize Highly Decorated Xylans.
J.Biol.Chem., 291, 2016
|
|
5NGL
 
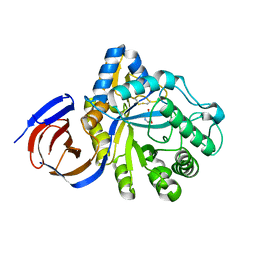 | | The endo-beta1,6-glucanase BT3312 | | Descriptor: | 1-DEOXYNOJIRIMYCIN, Glucosylceramidase, SODIUM ION, ... | | Authors: | Basle, A, Temple, M, Cuskin, F, Lowe, E, Gilbert, H. | | Deposit date: | 2017-03-17 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Bacteroidetes locus dedicated to fungal 1,6-beta-glucan degradation: Unique substrate conformation drives specificity of the key endo-1,6-beta-glucanase.
J. Biol. Chem., 292, 2017
|
|
4AZZ
 
 | |
5G5D
 
 | | Crystal Structure of the CohScaC2-XDocCipA type II complex from Clostridium thermocellum | | Descriptor: | CALCIUM ION, CELLULOSOMAL-SCAFFOLDING PROTEIN A, CELLULOSOME ANCHORING PROTEIN COHESIN REGION | | Authors: | Carvalho, A.L, A Bras, J.L, Najmudin, S.H, Pinheiro, B.A, Fontes, C.M.G.A. | | Deposit date: | 2016-05-23 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Diverse specificity of cellulosome attachment to the bacterial cell surface.
Sci Rep, 6, 2016
|
|
5G56
 
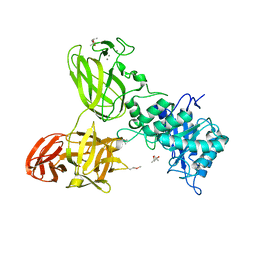 | | THE TETRA-MODULAR CELLULOSOMAL ARABINOXYLANASE CtXyl5A STRUCTURE AS REVEALED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CARBOHYDRATE BINDING FAMILY 6 | | Authors: | Bras, J.L.A, Gilbert, H.J, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2016-05-21 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The Mechanism by which Arabinoxylanases Can Recognise Highly Decorated Xylans.
J.Biol.Chem., 291, 2016
|
|
5JTS
 
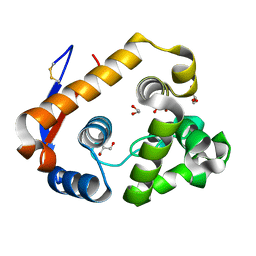 | | Structure of a beta-1,4-mannanase, SsGH134. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Jin, Y, Petricevic, M, Goddard-Borger, E.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2016-05-09 | | Release date: | 2016-11-16 | | Last modified: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | A beta-Mannanase with a Lysozyme-like Fold and a Novel Molecular Catalytic Mechanism.
ACS Cent Sci, 2, 2016
|
|
6G09
 
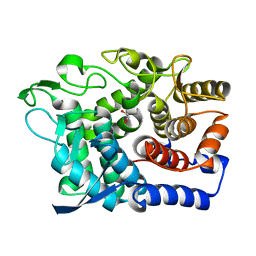 | | Crystal Structure of a GH8 xylobiose complex from Teredinibacter turnerae | | Descriptor: | 1,2-ETHANEDIOL, Glycoside hydrolase family 8 domain protein, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Fowler, C.A, Davies, G.J, Walton, P.H. | | Deposit date: | 2018-03-16 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and function of a glycoside hydrolase family 8 endoxylanase from Teredinibacter turnerae.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6G0B
 
 | |
6G00
 
 | | Crystal Structure of a GH8 xylanase from Teredinibacter turnerae | | Descriptor: | 1,2-ETHANEDIOL, Glycoside hydrolase family 8 domain protein, SODIUM ION | | Authors: | Fowler, C.A, Davies, G.J, Walton, P.H. | | Deposit date: | 2018-03-15 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and function of a glycoside hydrolase family 8 endoxylanase from Teredinibacter turnerae.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6G0N
 
 | | Crystal Structure of a GH8 catalytic mutant xylohexaose complex xylanase from Teredinibacter turnerae | | Descriptor: | GLYCEROL, Glycoside hydrolase family 8 domain protein, beta-D-xylopyranose, ... | | Authors: | Fowler, C.A, Davies, G.J, Walton, P.H. | | Deposit date: | 2018-03-19 | | Release date: | 2018-10-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and function of a glycoside hydrolase family 8 endoxylanase from Teredinibacter turnerae.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5JUG
 
 | | Structure of an inactive (E45Q) variant of a beta-1,4-mannanase, SsGH134, in complex with Man5 | | Descriptor: | CHLORIDE ION, GLYCEROL, alpha-D-mannopyranose, ... | | Authors: | Jin, Y, Petricevic, M, Goddard-Borger, E.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2016-05-10 | | Release date: | 2016-11-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | A beta-Mannanase with a Lysozyme-like Fold and a Novel Molecular Catalytic Mechanism.
ACS Cent Sci, 2, 2016
|
|
5JU9
 
 | | Structure of a beta-1,4-mannanase, SsGH134, in complex with Man3. | | Descriptor: | CHLORIDE ION, beta-1,4-mannanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Jin, Y, Petricevic, M, Goddard-Borger, E.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2016-05-10 | | Release date: | 2016-11-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | A beta-Mannanase with a Lysozyme-like Fold and a Novel Molecular Catalytic Mechanism.
ACS Cent Sci, 2, 2016
|
|
5K39
 
 | | THE TYPE II COHESIN DOCKERIN COMPLEX FROM CLOSTRIDIUM THERMOCELLUM | | Descriptor: | CALCIUM ION, Cellulosome anchoring protein cohesin region, Dockerin module from a protein of unknown function | | Authors: | Viegas, A, Pinheiro, B, Bras, J.L.A, Romao, M.J, Alves, V, Carvalho, A.L, Fontes, C.M.G.A. | | Deposit date: | 2016-05-19 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Diverse specificity of cellulosome attachment to the bacterial cell surface.
Sci Rep, 6, 2016
|
|
5M0Y
 
 | | Crystal Structure of the CohScaA-XDocCipB type II complex from Clostridium thermocellum at 1.5Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cellulosome anchoring protein cohesin region, ... | | Authors: | Pinheiro, B.A, Bras, J.L, Carvalho, A.L, Fontes, C.M.G.A. | | Deposit date: | 2016-10-06 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Diverse specificity of cellulosome attachment to the bacterial cell surface.
Sci Rep, 6, 2016
|
|
