2JG5
 
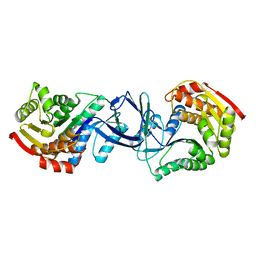 | | CRYSTAL STRUCTURE OF A PUTATIVE PHOSPHOFRUCTOKINASE FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | FRUCTOSE 1-PHOSPHATE KINASE | | Authors: | Yan, X, Carter, L.G, Johnson, K.A, Liu, H, Dorward, M, McMahon, S.A, Oke, M, Powers, H, Coote, P.J, Naismith, J.H. | | Deposit date: | 2007-02-08 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2JG6
 
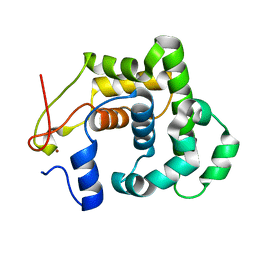 | | CRYSTAL STRUCTURE OF A 3-METHYLADENINE DNA GLYCOSYLASE I FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | DNA-3-METHYLADENINE GLYCOSIDASE, ZINC ION | | Authors: | Yan, X, Carter, L.G, Liu, H, Dorward, M, McMahon, S.A, Johnson, K.A, Oke, M, Coote, P.J, Naismith, J.H. | | Deposit date: | 2007-02-08 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X5T
 
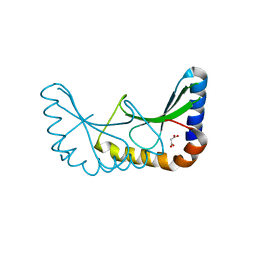 | | Crystal structure of ORF131 from Sulfolobus islandicus rudivirus 1 | | Descriptor: | MALONATE ION, ORF 131 | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, Naismith, J.H, White, M.F. | | Deposit date: | 2010-02-10 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X7B
 
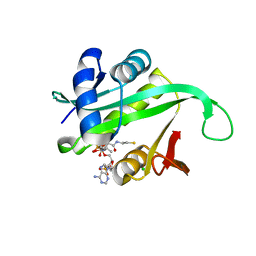 | | Crystal structure of the N-terminal acetylase Ard1 from Sulfolobus solfataricus P2 | | Descriptor: | CHLORIDE ION, COENZYME A, N-ACETYLTRANSFERASE SSO0209 | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, Mackay, D, White, M.F, Taylor, G.L, Naismith, J.H. | | Deposit date: | 2010-02-25 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X5R
 
 | | Crystal Structure of the hypothetical protein ORF126 from Pyrobaculum spherical virus | | Descriptor: | HYPOTHETICAL PROTEIN ORF126, ZINC ION | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-02-10 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X4K
 
 | | Crystal structure of SAR1376, a putative 4-oxalocrotonate tautomerase from the methicillin-resistant Staphylococcus aureus (MRSA) | | Descriptor: | 4-OXALOCROTONATE TAUTOMERASE, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-02-01 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X3N
 
 | | Crystal structure of pqsL, a probable FAD-dependent monooxygenase from Pseudomonas aeruginosa | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROBABLE FAD-DEPENDENT MONOOXYGENASE | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-25 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X4H
 
 | | Crystal Structure of the hypothetical protein SSo2273 from Sulfolobus solfataricus | | Descriptor: | HYPOTHETICAL PROTEIN SSO2273, ZINC ION | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-31 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2XU2
 
 | | Crystal Structure of the hypothetical protein PA4511 from Pseudomonas aeruginosa | | Descriptor: | CITRIC ACID, UPF0271 PROTEIN PA4511 | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, McMahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-10-14 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X5F
 
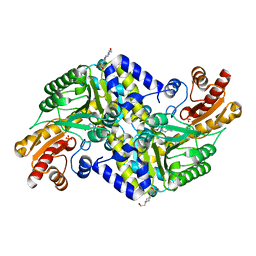 | | Crystal structure of the methicillin-resistant Staphylococcus aureus Sar2028, an aspartate_tyrosine_phenylalanine pyridoxal-5'-phosphate dependent aminotransferase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ASPARTATE_TYROSINE_PHENYLALANINE PYRIDOXAL-5' PHOSPHATE-DEPENDENT AMINOTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-02-08 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genom., 11, 2010
|
|
2X3F
 
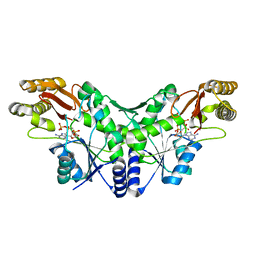 | | Crystal Structure of the Methicillin-Resistant Staphylococcus aureus Sar2676, a Pantothenate Synthetase. | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, PANTHOTHENATE SYNTHETASE, SULFATE ION | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-24 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genom., 11, 2010
|
|
2IVY
 
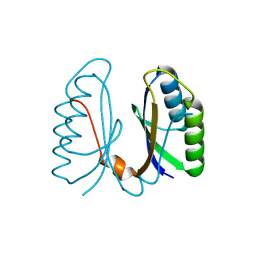 | | Crystal structure of hypothetical protein sso1404 from Sulfolobus solfataricus P2 | | Descriptor: | HYPOTHETICAL PROTEIN SSO1404 | | Authors: | Yan, X, Carter, L.G, Dorward, M, Liu, H, McMahon, S.A, Oke, M, Powers, H, White, M.F, Naismith, J.H. | | Deposit date: | 2006-06-22 | | Release date: | 2006-06-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
4AI4
 
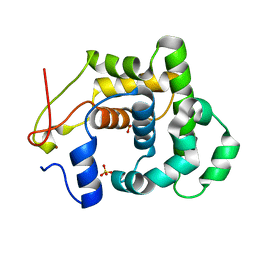 | |
4AI5
 
 | |
4AIA
 
 | |
2VW8
 
 | | Crystal Structure of Quinolone signal response protein pqsE from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, FE (II) ION, ... | | Authors: | Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, Oke, M, Naismith, J.H, White, M.F. | | Deposit date: | 2008-06-17 | | Release date: | 2010-07-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2VXZ
 
 | | Crystal Structure of hypothetical protein PyrSV_gp04 from Pyrobaculum spherical virus | | Descriptor: | CHLORIDE ION, GLYCEROL, PYRSV_GP04 | | Authors: | Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, Oke, M, Naismith, J.H, White, M.F. | | Deposit date: | 2008-07-15 | | Release date: | 2009-11-17 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2WJ9
 
 | | ArdB | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Weikart, N.D, Roberts, G, Johnson, K.A, Oke, M, Cooper, L.P, McMahon, S.A, White, J.H, Liu, H, Carter, L.G, Walkinshaw, M.D, Blakely, G.W, Naismith, J.H, Dryden, D.T.F. | | Deposit date: | 2009-05-25 | | Release date: | 2010-08-18 | | Last modified: | 2018-05-02 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X48
 
 | | ORF 55 from Sulfolobus islandicus rudivirus 1 | | Descriptor: | CAG38821, PHOSPHATE ION | | Authors: | Oke, M, Carter, L, Johnson, K.A, Liu, H, Mcmahon, S, Naismith, J.H, White, M.F. | | Deposit date: | 2010-01-28 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X0O
 
 | | Apo structure of the Alcaligin biosynthesis protein C (AlcC) from Bordetella bronchiseptica | | Descriptor: | ALCALIGIN BIOSYNTHESIS PROTEIN, SULFATE ION | | Authors: | Johnson, K.A, Schmelz, S, Kadi, N, Mcmahon, S.A, Oke, M, Liu, H, Carter, L.G, White, M.F, Challis, G.L, Naismith, J.H. | | Deposit date: | 2009-12-16 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X4J
 
 | | Crystal structure of ORF137 from Pyrobaculum spherical virus | | Descriptor: | HYPOTHETICAL PROTEIN ORF137 | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-02-01 | | Release date: | 2010-07-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X3E
 
 | | Crystal structure of 3-oxoacyl-(acyl carrier protein) synthase III, FabH from Pseudomonas aeruginosa PAO1 | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] SYNTHASE 3 | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-24 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X3L
 
 | | Crystal Structure of the Orn_Lys_Arg decarboxylase family protein SAR0482 from Methicillin-resistant Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, ORN/LYS/ARG DECARBOXYLASE FAMILY PROTEIN, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-25 | | Release date: | 2010-07-21 | | Last modified: | 2015-11-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X5C
 
 | | Crystal structure of hypothetical protein ORF131 from Pyrobaculum Spherical Virus | | Descriptor: | GLYCEROL, HYPOTHETICAL PROTEIN ORF131, ZINC ION | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-02-08 | | Release date: | 2010-07-21 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genom., 11, 2010
|
|
2X4G
 
 | | Crystal structure of PA4631, a nucleoside-diphosphate-sugar epimerase from Pseudomonas aeruginosa | | Descriptor: | NUCLEOSIDE-DIPHOSPHATE-SUGAR EPIMERASE | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-30 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
