6MD5
 
 | |
1MGQ
 
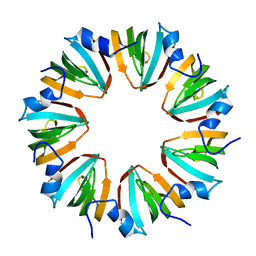 | |
1I81
 
 | | CRYSTAL STRUCTURE OF A HEPTAMERIC LSM PROTEIN FROM METHANOBACTERIUM THERMOAUTOTROPHICUM | | Descriptor: | PUTATIVE SNRNP SM-LIKE PROTEIN | | Authors: | Collins, B.M, Harrop, S.J, Kornfeld, G.D, Dawes, I.W, Curmi, P.M.G, Mabbutt, B.C. | | Deposit date: | 2001-03-12 | | Release date: | 2001-03-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a heptameric Sm-like protein complex from archaea: implications for the structure and evolution of snRNPs.
J.Mol.Biol., 309, 2001
|
|
1Z2X
 
 | | Crystal structure of mouse Vps29 | | Descriptor: | Vacuolar protein sorting 29 | | Authors: | Collins, B.M, Skinner, C.F, Watson, P.J, Seaman, M.N.J, Owen, D.J. | | Deposit date: | 2005-03-10 | | Release date: | 2005-06-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Vps29 has a phosphoesterase fold that acts as a protein interaction scaffold for retromer assembly
NAT.STRUCT.MOL.BIOL., 12, 2005
|
|
1Z2W
 
 | | Crystal structure of mouse Vps29 complexed with Mn2+ | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Vacuolar protein sorting 29 | | Authors: | Collins, B.M, Skinner, C.F, Watson, P.J, Seaman, M.N.J, Owen, D.J. | | Deposit date: | 2005-03-10 | | Release date: | 2005-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Vps29 has a phosphoesterase fold that acts as a protein interaction scaffold for retromer assembly
NAT.STRUCT.MOL.BIOL., 12, 2005
|
|
1N9S
 
 | | Crystal structure of yeast SmF in spacegroup P43212 | | Descriptor: | Small nuclear ribonucleoprotein F | | Authors: | Collins, B.M, Cubeddu, L, Naidoo, N, Harrop, S.J, Kornfeld, G.D, Dawes, I.W, Curmi, P.M.G, Mabbutt, B.C. | | Deposit date: | 2002-11-26 | | Release date: | 2002-12-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Homomeric ring assemblies of eukaryotic Sm proteins have affinity for
both RNA and DNA: Crystal structure of an oligomeric complex of yeast
SmF
J.Biol.Chem., 278, 2003
|
|
1N9R
 
 | | Crystal structure of a heptameric ring complex of yeast SmF in spacegroup P4122 | | Descriptor: | Small nuclear ribonucleoprotein F | | Authors: | Collins, B.M, Cubeddu, L, Naidoo, N, Harrop, S.J, Kornfeld, G.D, Dawes, I.W, Curmi, P.M.G, Mabbutt, B.C. | | Deposit date: | 2002-11-26 | | Release date: | 2002-12-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Homomeric ring assemblies of eukaryotic Sm proteins have affinity for
both RNA and DNA: Crystal structure of an oligomeric complex of yeast
SmF
J.Biol.Chem., 278, 2003
|
|
1NAF
 
 | |
1OM9
 
 | | Structure of the GGA1-appendage in complex with the p56 binding peptide | | Descriptor: | 15-mer peptide fragment of p56, ADP-ribosylation factor binding protein GGA1 | | Authors: | Collins, B.M, Praefcke, G.J.K, Robinson, M.S, Owen, D.J. | | Deposit date: | 2003-02-25 | | Release date: | 2003-07-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for binding of accessory proteins by the appendage domain of GGAs
Nat.Struct.Biol., 10, 2003
|
|
7SR2
 
 | | Crystal structure of the human SNX25 regulator of G-protein signalling (RGS) domain | | Descriptor: | ACETATE ION, LEUCINE, Sorting nexin-25, ... | | Authors: | Collins, B.M, Paul, B, Weeratunga, S. | | Deposit date: | 2021-11-07 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Predictions of the SNX-RGS Proteins Suggest They Belong to a New Class of Lipid Transfer Proteins.
Front Cell Dev Biol, 10, 2022
|
|
7SR1
 
 | |
5W8M
 
 | | Crystal structure of Chaetomium thermophilum Vps29 | | Descriptor: | GLYCEROL, TRIETHYLENE GLYCOL, Vacuolar protein sorting-associated protein 29 | | Authors: | Collins, B.M, Leneva, N. | | Deposit date: | 2017-06-21 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure of the membrane-assembled retromer coat determined by cryo-electron tomography.
Nature, 561, 2018
|
|
4P2I
 
 | | Crystal structure of the mouse SNX19 PX domain | | Descriptor: | MKIAA0254 protein | | Authors: | Collins, B.M. | | Deposit date: | 2014-03-04 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Different Phosphoinositide Specificities of the PX Domains of Sorting Nexins Regulating G-protein Signaling.
J.Biol.Chem., 289, 2014
|
|
4P2J
 
 | |
5ELQ
 
 | |
5EM9
 
 | |
5EMA
 
 | |
5EMB
 
 | |
2R51
 
 | | Crystal Structure of mouse Vps26B | | Descriptor: | Vacuolar protein sorting-associated protein 26B | | Authors: | Owen, D.J, Teasdale, R.D, Collins, B.M. | | Deposit date: | 2007-09-02 | | Release date: | 2008-07-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Vps26B and mapping of its interaction with the retromer protein complex.
Traffic, 9, 2008
|
|
2VGL
 
 | | AP2 CLATHRIN ADAPTOR CORE | | Descriptor: | ADAPTOR PROTEIN COMPLEX AP-2, ALPHA 2 SUBUNIT, AP-2 COMPLEX SUBUNIT BETA-1, ... | | Authors: | Owen, D.J, Collins, B.M, McCoy, A.J, Evans, P.R. | | Deposit date: | 2007-11-14 | | Release date: | 2007-12-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Architecture and Functional Model of the Endocytic Ap2 Complex
Cell(Cambridge,Mass.), 109, 2002
|
|
5WOE
 
 | |
6MBI
 
 | |
7JJC
 
 | |
6XS7
 
 | | Crystal structure of human Vps29 complexed with RaPID-derived cyclic peptide RT-D2 | | Descriptor: | 48V-DTY-THR-THR-ILE-TYR-TRP-THR-PRO-LEU-GLY-THR-PHE-PRO-ARG-ILE-ARG, FORMIC ACID, GLYCEROL, ... | | Authors: | Chen, K.-E, Guo, Q, Collins, B.M. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | De novo macrocyclic peptides for inhibiting, stabilizing, and probing the function of the retromer endosomal trafficking complex.
Sci Adv, 7, 2021
|
|
6XSA
 
 | | Crystal structure of human Vps29 complexed with RaPID-derived cyclic peptide RT-L2 | | Descriptor: | 48V-TYR-LEU-PRO-THR-ILE-THR-GLY-VAL-GLY-HIS-LEU-TRP-HIS-PRO-LEU, SULFATE ION, Vacuolar protein sorting-associated protein 29 | | Authors: | Chen, K.-E, Guo, Q, Collins, B.M. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | De novo macrocyclic peptides for inhibiting, stabilizing, and probing the function of the retromer endosomal trafficking complex.
Sci Adv, 7, 2021
|
|
