3LA7
 
 | | Crystal structure of NtcA in apo-form | | Descriptor: | Global nitrogen regulator, octyl beta-D-glucopyranoside | | Authors: | Zhao, M.X, Jiang, Y.L, He, Y.X, Chen, Y.F, Teng, Y.B, Zhang, C.C, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2010-01-06 | | Release date: | 2010-09-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the allosteric control of the global transcription factor NtcA by the nitrogen starvation signal 2-oxoglutarate.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3LA2
 
 | | Crystal structure of NtcA in complex with 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, Global nitrogen regulator | | Authors: | Zhao, M.X, Jiang, Y.L, He, Y.X, Chen, Y.F, Teng, Y.B, Chen, Y.X, Zhang, C.C, Zhou, C.Z. | | Deposit date: | 2010-01-06 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the allosteric control of the global transcription factor NtcA by the nitrogen starvation signal 2-oxoglutarate.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3LA3
 
 | | Crystal structure of NtcA in complex with 2,2-difluoropentanedioic acid | | Descriptor: | 2,2-difluoropentanedioic acid, Global nitrogen regulator | | Authors: | Zhao, M.X, Jiang, Y.L, He, Y.X, Chen, Y.F, Teng, Y.B, Chen, Y.X, Zhang, C.C, Zhou, C.Z. | | Deposit date: | 2010-01-06 | | Release date: | 2010-07-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the allosteric control of the global transcription factor NtcA by the nitrogen starvation signal 2-oxoglutarate.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MRS
 
 | |
6INR
 
 | | The crystal structure of phytoplasmal effector causing phyllody symptoms 1 (PHYL1) | | Descriptor: | CADMIUM ION, Putative effector, AYWB SAP54-like protein | | Authors: | Liao, Y.T, Lin, S.S, Ko, T.P, Wang, H.C. | | Deposit date: | 2018-10-26 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural insights into the interaction between phytoplasmal effector causing phyllody 1 and MADS transcription factors.
Plant J., 100, 2019
|
|
3N2E
 
 | | Crystal structure of Helicobactor pylori shikimate kinase in complex with NSC162535 | | Descriptor: | 7-amino-4-hydroxy-3-[(E)-(5-hydroxy-7-sulfonaphthalen-2-yl)diazenyl]naphthalene-2-sulfonic acid, L(+)-TARTARIC ACID, Shikimate kinase | | Authors: | Cheng, W.C, Chen, T.J, Lin, S.C, Wang, W.C. | | Deposit date: | 2010-05-18 | | Release date: | 2011-05-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structures of Helicobacter pylori shikimate kinase reveal a selective inhibitor-induced-fit mechanism
Plos One, 7, 2012
|
|
3MUF
 
 | | Shikimate kinase from Helicobacter pylori in complex with shikimate-3-phosphate and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SHIKIMATE-3-PHOSPHATE, Shikimate kinase | | Authors: | Cheng, W.C, Chen, T.J, Lin, S.C, Wang, W.C. | | Deposit date: | 2010-05-03 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Helicobacter pylori shikimate kinase reveal a selective inhibitor-induced-fit mechanism
Plos One, 7, 2012
|
|
3HR7
 
 | |
6NSV
 
 | | Crystal structure of the human CHIP TPR domain in complex with a 5mer acetylated optimized peptide | | Descriptor: | ACE-LEU-TRP-TRP-PRO-ASP, CHLORIDE ION, E3 ubiquitin-protein ligase CHIP, ... | | Authors: | Basu, K, Ravalin, M, Bohn, M.-F, Craik, C.S, Gestwicki, J.E. | | Deposit date: | 2019-01-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.305 Å) | | Cite: | Specificity for latent C termini links the E3 ubiquitin ligase CHIP to caspases.
Nat.Chem.Biol., 15, 2019
|
|
6EFK
 
 | | Crystal structure of the human CHIP TPR domain in complex with a 5mer acetylated HSP70 peptide | | Descriptor: | ACE-ILE-GLU-GLU-VAL-ASP, E3 ubiquitin-protein ligase CHIP, SODIUM ION | | Authors: | Basu, K, Ravalin, M, Bohn, M.-F, Craik, C.S, Gestwicki, J.E. | | Deposit date: | 2018-08-16 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Specificity for latent C termini links the E3 ubiquitin ligase CHIP to caspases.
Nat.Chem.Biol., 15, 2019
|
|
7T32
 
 | | CryoEM structure of the adenosine 2A receptor-BRIL/Anti BRIL Fab complex with ZM241385 | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a/Soluble cytochrome b562 Fusion Protein | | Authors: | Zhang, K.H, Wu, H, Hoppe, N, Manglik, A, Cheng, Y.F. | | Deposit date: | 2021-12-06 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Fusion protein strategies for cryo-EM study of G protein-coupled receptors.
Nat Commun, 13, 2022
|
|
8EUF
 
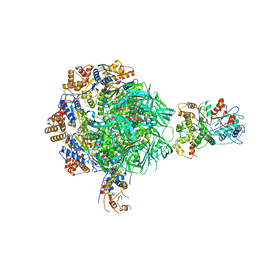 | | Class2 of the INO80-Nucleosome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling ATPase INO80, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Narlikar, G, Cheng, Y.F. | | Deposit date: | 2022-10-18 | | Release date: | 2023-07-12 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8EUJ
 
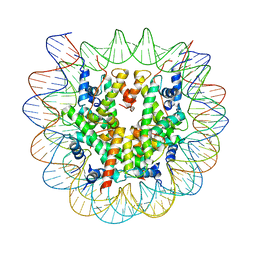 | | Class2 of the INO80-Nucleosome complex | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Narlikar, G, Cheng, Y.F. | | Deposit date: | 2022-10-18 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8ETT
 
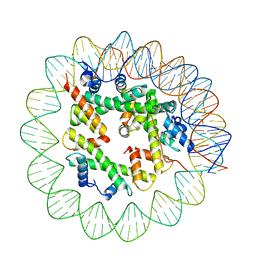 | | Class1 of the INO80-Hexasome complex | | Descriptor: | DNA (110-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Cheng, Y.F, Narlikar, G. | | Deposit date: | 2022-10-17 | | Release date: | 2023-07-12 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (6.68 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8ETV
 
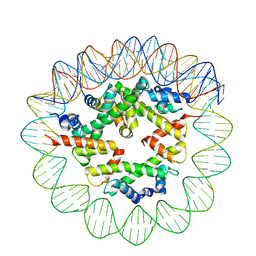 | | Class2 of the INO80-Hexasome complex | | Descriptor: | DNA (110-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Cheng, Y.F, Narlikar, G. | | Deposit date: | 2022-10-17 | | Release date: | 2023-07-12 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8EU2
 
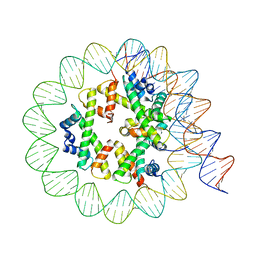 | | Class3 of the INO80-Hexasome complex | | Descriptor: | DNA (110-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Cheng, Y.F, Narlikar, G. | | Deposit date: | 2022-10-18 | | Release date: | 2023-07-12 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8EUE
 
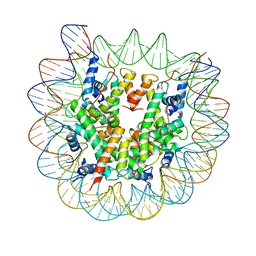 | | Class1 of the INO80-Nucleosome complex | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Narlikar, G, Cheng, Y.F. | | Deposit date: | 2022-10-18 | | Release date: | 2023-07-12 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8EU9
 
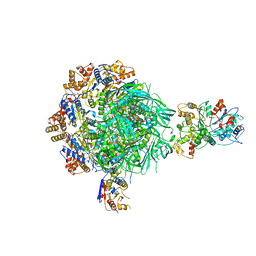 | | Class1 of the INO80-Nucleosome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling ATPase INO80, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Cheng, Y.F, Narlikar, G. | | Deposit date: | 2022-10-18 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8ETS
 
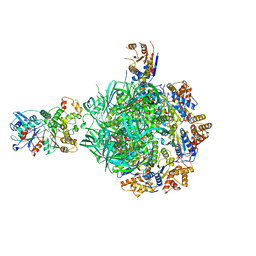 | | Class1 of the INO80-Hexasome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling ATPase INO80, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Cheng, Y.F, Narlikar, G. | | Deposit date: | 2022-10-17 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8ETU
 
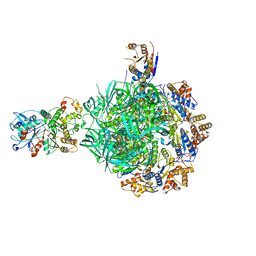 | | Class2 of the INO80-Hexasome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling ATPase INO80, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Cheng, Y.F, Narlikar, G. | | Deposit date: | 2022-10-17 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8ETW
 
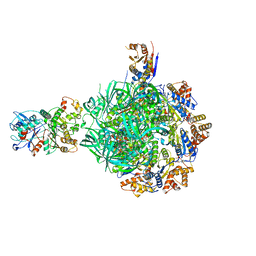 | | Class3 of INO80-Hexasome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling ATPase INO80, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Narlikar, G, Cheng, Y.F. | | Deposit date: | 2022-10-17 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8G17
 
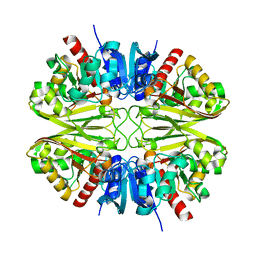 | | CryoEM structure of wild-type GAPDH | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Choi, W.Y, Wu, H, Cheng, Y.F. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (1.98 Å) | | Cite: | Efficient tagging of endogenous proteins in human cell lines for structural studies by single-particle cryo-EM.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8G13
 
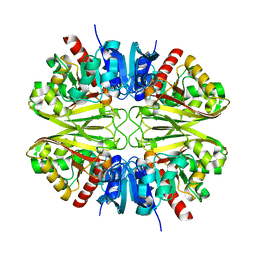 | |
8G12
 
 | |
8G16
 
 | |
