1LIA
 
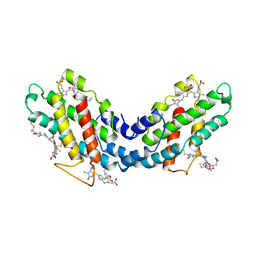 | |
2NSF
 
 | | Crystal structure of the mycothiol-dependent maleylpyruvate isomerase | | Descriptor: | GLYCEROL, Hypothetical protein Cgl3021, SULFATE ION, ... | | Authors: | Chang, W.R, Wang, R. | | Deposit date: | 2006-11-04 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structures and Site-directed Mutagenesis of a Mycothiol-dependent Enzyme Reveal a Novel Folding and Molecular Basis for Mycothiol-mediated Maleylpyruvate Isomerization
J.Biol.Chem., 282, 2007
|
|
3RE1
 
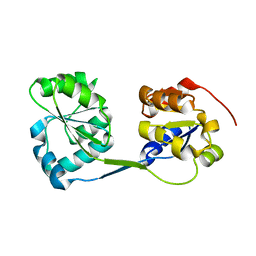 | |
1JYB
 
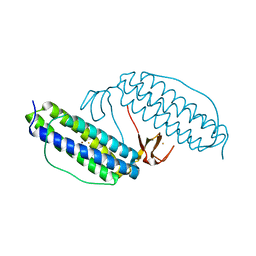 | | Crystal structure of Rubrerythrin | | Descriptor: | FE (III) ION, Rubrerythrin, ZINC ION | | Authors: | Chang, W.R, Li, M, Liu, M.Y. | | Deposit date: | 2001-09-11 | | Release date: | 2002-09-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure studies on rubrerythrin: enzymatic activity in relation to the zinc movement.
J.Biol.Inorg.Chem., 8, 2003
|
|
2NSG
 
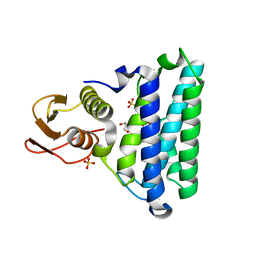 | |
3JCU
 
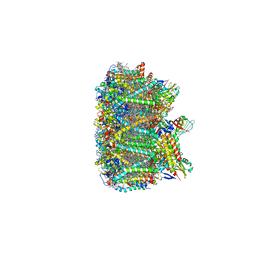 | | Cryo-EM structure of spinach PSII-LHCII supercomplex at 3.2 Angstrom resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Wei, X.P, Zhang, X.Z, Su, X.D, Cao, P, Liu, X.Y, Li, M, Chang, W.R, Liu, Z.F. | | Deposit date: | 2016-03-10 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of spinach photosystem II-LHCII supercomplex at 3.2 A resolution
Nature, 534, 2016
|
|
3K0T
 
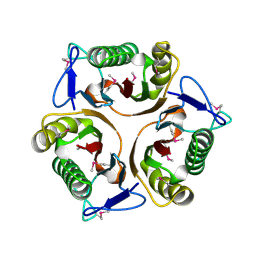 | |
2AVF
 
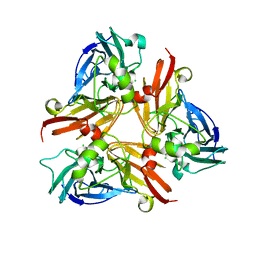 | | Crystal Structure of C-terminal Desundecapeptide Nitrite Reductase from Achromobacter cycloclastes | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Li, H.T, Chang, T, Chang, W.C, Chen, C.J, Liu, M.Y, Gui, L.L, Zhang, J.P, An, X.M, Chang, W.R. | | Deposit date: | 2005-08-30 | | Release date: | 2005-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of C-terminal desundecapeptide nitrite reductase from Achromobacter cycloclastes
Biochem.Biophys.Res.Commun., 338, 2005
|
|
1KCB
 
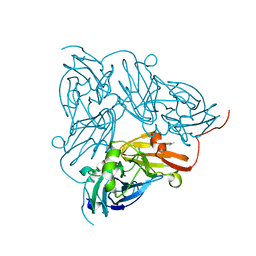 | | Crystal Structure of a NO-forming Nitrite Reductase Mutant: an Analog of a Transition State in Enzymatic Reaction | | Descriptor: | COPPER (II) ION, Nitrite Reductase | | Authors: | Liu, S.Q, Chang, T, Liu, M.Y, LeGall, J, Chang, W.C, Zhang, J.P, Liang, D.C, Chang, W.R. | | Deposit date: | 2001-11-07 | | Release date: | 2003-11-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a NO-forming nitrite reductase mutant: an analog of a transition state in enzymatic reaction
Biochem.Biophys.Res.Commun., 302, 2003
|
|
1RZQ
 
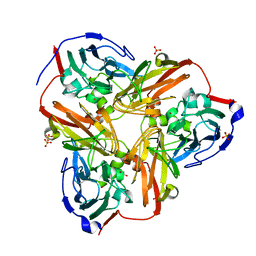 | | Crystal Structure of C-Terminal Despentapeptide Nitrite Reductase from Achromobacter Cycloclastes at pH5.0 | | Descriptor: | ACETIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Li, H.T, Wang, C, Chang, T, Chang, W.C, Liu, M.Y, Le Gall, J, Gui, L.L, Zhang, J.P, An, X.M, Chang, W.R. | | Deposit date: | 2003-12-26 | | Release date: | 2004-03-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | pH-profile crystal structure studies of C-terminal despentapeptide nitrite reductase from Achromobacter cycloclastes
Biochem.Biophys.Res.Commun., 316, 2004
|
|
1RZP
 
 | | Crystal Structure of C-Terminal Despentapeptide Nitrite Reductase from Achromobacter Cycloclastes at pH6.2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Li, H.T, Wang, C, Chang, T, Chang, W.C, Liu, M.Y, Le Gall, J, Gui, L.L, Zhang, J.P, An, X.M, Chang, W.R. | | Deposit date: | 2003-12-26 | | Release date: | 2004-03-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | pH-profile crystal structure studies of C-terminal despentapeptide nitrite reductase from Achromobacter cycloclastes
Biochem.Biophys.Res.Commun., 316, 2004
|
|
1JR9
 
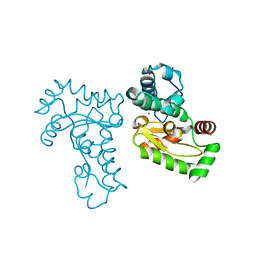 | | Crystal Structure of manganese superoxide dismutases from Bacillus halodenitrificans | | Descriptor: | MANGANESE (II) ION, ZINC ION, manganese superoxide dismutase | | Authors: | Liao, J, Liu, M.Y, Chang, T, Li, M, LeGall, J, Gui, L.L, Zhang, J.P, Jiang, T, Liang, D.C, Chang, W.R. | | Deposit date: | 2001-08-13 | | Release date: | 2002-08-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of manganese superoxide dismutase from Bacillus halodenitrificans, a component of the so-called "green protein".
J.Struct.Biol., 139, 2002
|
|
3RDY
 
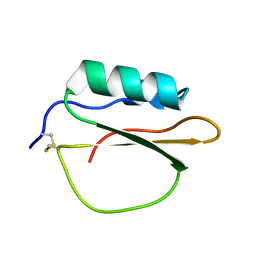 | |
3RDZ
 
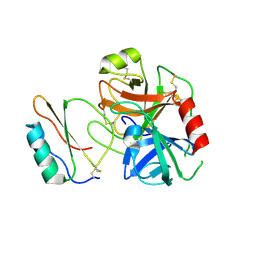 | |
1YM0
 
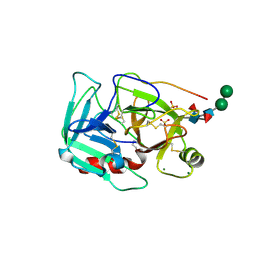 | | Crystal Structure of Earthworm Fibrinolytic Enzyme Component B: a Novel, Glycosylated Two-chained Trypsin | | Descriptor: | MAGNESIUM ION, SULFATE ION, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, F, Wang, C, Li, M, Zhang, J.P, Gui, L.L, An, X.M, Chang, W.R. | | Deposit date: | 2005-01-20 | | Release date: | 2005-04-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of earthworm fibrinolytic enzyme component B: a novel, glycosylated two-chained trypsin.
J.Mol.Biol., 348, 2005
|
|
1F99
 
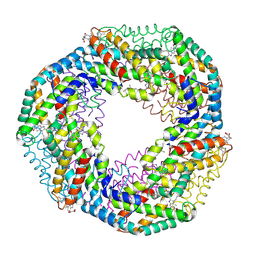 | |
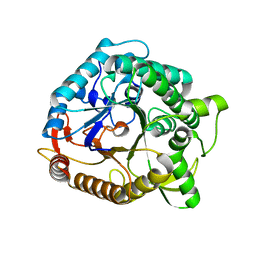
1NP2
 
 | |
2A3R
 
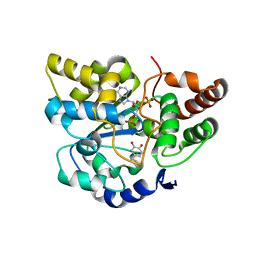 | | Crystal Structure of Human Sulfotransferase SULT1A3 in Complex with Dopamine and 3-Phosphoadenosine 5-Phosphate | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, L-DOPAMINE, Monoamine-sulfating phenol sulfotransferase | | Authors: | Lu, J.H, Li, H.T, Liu, M.C, Zhang, J.P, Li, M, An, X.M, Chang, W.R. | | Deposit date: | 2005-06-26 | | Release date: | 2005-08-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human sulfotransferase SULT1A3 in complex with dopamine and 3'-phosphoadenosine 5'-phosphate
Biochem.Biophys.Res.Commun., 335, 2005
|
|
1ZJ6
 
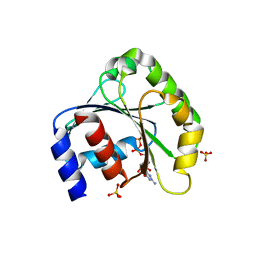 | | Crystal structure of human ARL5 | | Descriptor: | ADP-ribosylation factor-like protein 5, GUANOSINE-3'-MONOPHOSPHATE-5'-DIPHOSPHATE, SULFATE ION | | Authors: | Wang, Z.X, Shi, L, Liu, J.F, An, X.M, Chang, W.R, Liang, D.C. | | Deposit date: | 2005-04-28 | | Release date: | 2005-06-07 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0A crystal structure of human ARL5-GDP3'P, a novel member of the small GTP-binding proteins
Biochem.Biophys.Res.Commun., 332, 2005
|
|
1RFJ
 
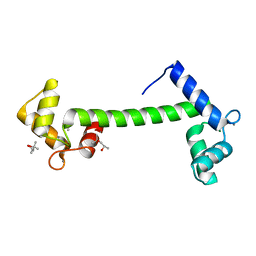 | | Crystal Structure of Potato Calmodulin PCM6 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, calmodulin | | Authors: | Liang, D.C, Yun, C.H, Chang, W.R. | | Deposit date: | 2003-11-10 | | Release date: | 2004-06-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of potato calmodulin PCM6: the first report of the three-dimensional structure of a plant calmodulin.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1TGR
 
 | | Crystal Structure of mini-IGF-1(2) | | Descriptor: | Insulin-like growth factor IA | | Authors: | Liang, D.C, Yun, C.H, Chang, W.R. | | Deposit date: | 2004-05-29 | | Release date: | 2004-12-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | 1.42A crystal structure of mini-IGF-1(2): an analysis of the disulfide isomerization property and receptor binding property of IGF-1 based on the three-dimensional structure
Biochem.Biophys.Res.Commun., 326, 2004
|
|
3BXV
 
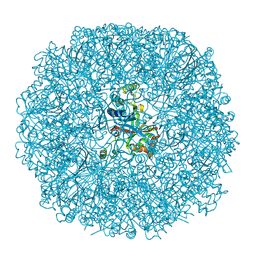 | |
1LHP
 
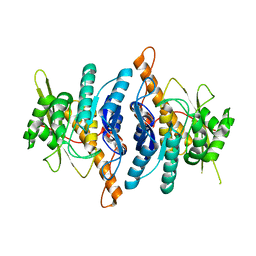 | |
5XNM
 
 | | Structure of unstacked C2S2M2-type PSII-LHCII supercomplex from Pisum sativum | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Su, X.D, Ma, J, Wei, X.P, Cao, P, Zhu, D.J, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2017-05-23 | | Release date: | 2017-09-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex
Science, 357, 2017
|
|
5XNL
 
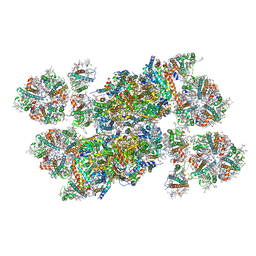 | | Structure of stacked C2S2M2-type PSII-LHCII supercomplex from Pisum sativum | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Su, X.D, Ma, J, Wei, X.P, Cao, P, Zhu, D.J, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2017-05-23 | | Release date: | 2017-09-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex
Science, 357, 2017
|
|
