3K1J
 
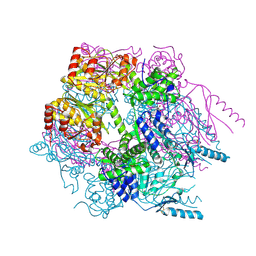 | | Crystal structure of Lon protease from Thermococcus onnurineus NA1 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Cha, S.S, An, Y.J. | | Deposit date: | 2009-09-28 | | Release date: | 2010-09-22 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Lon protease: molecular architecture of gated entry to a sequestered degradation chamber
Embo J., 29, 2010
|
|
3BWE
 
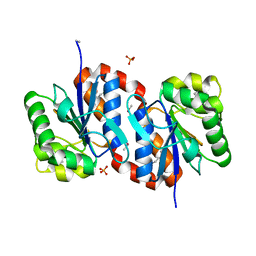 | | Crystal structure of aggregated form of DJ1 | | Descriptor: | PHOSPHATE ION, Protein DJ-1 | | Authors: | Cha, S.S. | | Deposit date: | 2008-01-09 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of filamentous aggregates of human DJ-1 formed in an inorganic phosphate-dependent manner.
J.Biol.Chem., 283, 2008
|
|
6XH1
 
 | |
6XH3
 
 | |
6XH2
 
 | |
6XH0
 
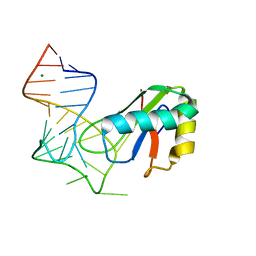 | |
8UZ1
 
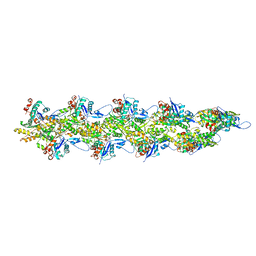 | | Straight actin filament from Arp2/3 branch junction sample (ADP-BeFx) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Chavali, S.S, Chou, S.Z, Sindelar, C.V. | | Deposit date: | 2023-11-14 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures reveal how phosphate release from Arp3 weakens actin filament branches formed by Arp2/3 complex.
Nat Commun, 15, 2024
|
|
8UXX
 
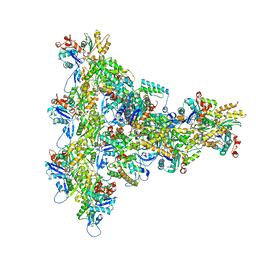 | | Arp2/3 branch junction complex, BeFx state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Chavali, S.S, Chou, S.Z, Sindelar, C.V. | | Deposit date: | 2023-11-11 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures reveal how phosphate release from Arp3 weakens actin filament branches formed by Arp2/3 complex.
Nat Commun, 15, 2024
|
|
8UXW
 
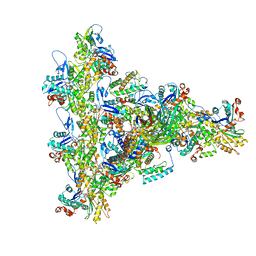 | | Arp2/3 branch junction complex, ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Chavali, S.S, Chou, S.Z, Sindelar, C.V. | | Deposit date: | 2023-11-11 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures reveal how phosphate release from Arp3 weakens actin filament branches formed by Arp2/3 complex.
Nat Commun, 15, 2024
|
|
8UZ0
 
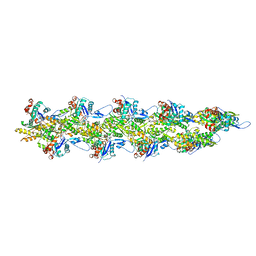 | | Straight actin filament from Arp2/3 branch junction sample (ADP) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Chavali, S.S, Chou, S.Z, Sindelar, C.V. | | Deposit date: | 2023-11-14 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures reveal how phosphate release from Arp3 weakens actin filament branches formed by Arp2/3 complex.
Nat Commun, 15, 2024
|
|
5GMX
 
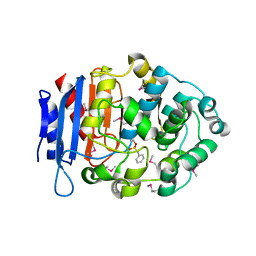 | | Crystal structure of a family VIII carboxylesterase | | Descriptor: | Carboxylesterase, phenylmethanesulfonic acid | | Authors: | An, Y.J, Cha, S.S. | | Deposit date: | 2016-07-18 | | Release date: | 2017-07-26 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of EstSRT1, a family VIII carboxylesterase displaying hydrolytic activity toward oxyimino cephalosporins
Biochem. Biophys. Res. Commun., 478, 2016
|
|
4QDI
 
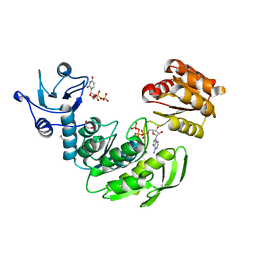 | | Crystal structure II of MurF from Acinetobacter baumannii | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | An, Y.J, Jeong, C.S, Cha, S.S. | | Deposit date: | 2014-05-13 | | Release date: | 2015-04-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ATP-binding mode including a carbamoylated lysine and two Mg(2+) ions, and substrate-binding mode in Acinetobacter baumannii MurF
Biochem.Biophys.Res.Commun., 450, 2014
|
|
4QF5
 
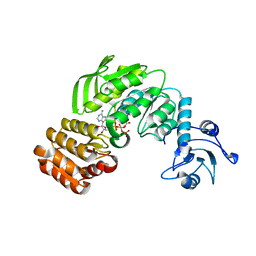 | | Crystal structure I of MurF from Acinetobacter baumannii | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, UDP-N-acetylmuramoyl-tripeptide--D-alanyl-D-alanine ligase | | Authors: | An, Y.J, Jeong, C.S, Cha, S.S. | | Deposit date: | 2014-05-19 | | Release date: | 2015-04-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | ATP-binding mode including a carbamoylated lysine and two Mg(2+) ions, and substrate-binding mode in Acinetobacter baumannii MurF
Biochem.Biophys.Res.Commun., 450, 2014
|
|
4DWZ
 
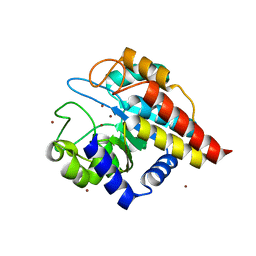 | | Crystal Structure of Ton_0340 | | Descriptor: | Hypothetical protein TON_0340, ZINC ION | | Authors: | Lee, S.G, Lee, K.H, Cha, S.S, Oh, B.H. | | Deposit date: | 2012-02-27 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Experimental phasing using zinc anomalous scattering
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DT3
 
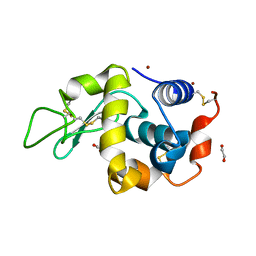 | | Crystal structure of zinc-charged lysozyme | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | An, Y.J, Jeong, C.S, Cha, S.S. | | Deposit date: | 2012-02-20 | | Release date: | 2012-09-12 | | Last modified: | 2013-07-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental phasing using zinc anomalous scattering
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4FC5
 
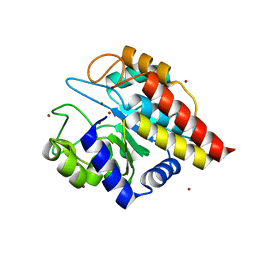 | | Crystal Structure of Ton_0340 | | Descriptor: | Putative uncharacterized protein, ZINC ION | | Authors: | Lee, S.G, Lee, K.H, An, Y.J, Cha, S.S, Oh, B.H. | | Deposit date: | 2012-05-24 | | Release date: | 2012-09-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Experimental phasing using zinc anomalous scattering
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3MVE
 
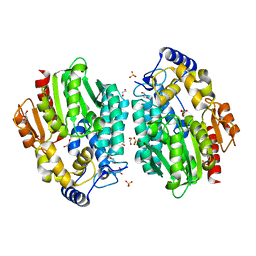 | | Crystal structure of a novel pyruvate decarboxylase | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, UPF0255 protein VV1_0328 | | Authors: | Cha, S.S, Jeong, C.S, An, Y.J. | | Deposit date: | 2010-05-04 | | Release date: | 2011-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | FrsA functions as a cofactor-independent decarboxylase to control metabolic flux.
Nat.Chem.Biol., 7, 2011
|
|
1IZZ
 
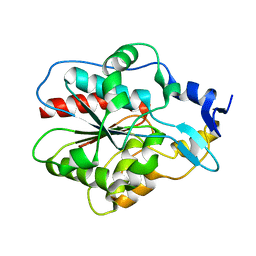 | | Crystal structure of Hsp31 | | Descriptor: | Hsp31 | | Authors: | Cha, S.S, Lee, S.J. | | Deposit date: | 2002-10-16 | | Release date: | 2003-10-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structures of human DJ-1 and Escherichia coli Hsp31, which share an evolutionarily conserved domain
J.Biol.Chem., 278, 2003
|
|
1IZY
 
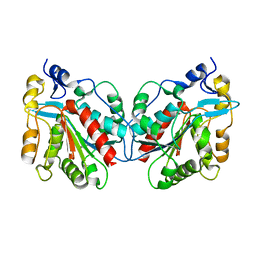 | | Crystal structure of Hsp31 | | Descriptor: | Hsp31 | | Authors: | Cha, S.S, Lee, S.J. | | Deposit date: | 2002-10-16 | | Release date: | 2003-10-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of human DJ-1 and Escherichia coli Hsp31, which share an evolutionarily conserved domain
J.Biol.Chem., 278, 2003
|
|
1J42
 
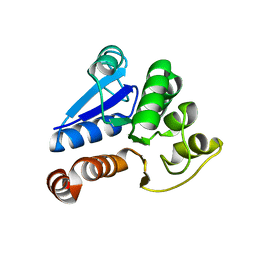 | | Crystal Structure of Human DJ-1 | | Descriptor: | RNA-binding protein regulatory subunit | | Authors: | Cha, S.S. | | Deposit date: | 2003-02-26 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of human DJ-1 and Escherichia coli Hsp31, which share an evolutionarily conserved domain.
J.Biol.Chem., 278, 2003
|
|
1K41
 
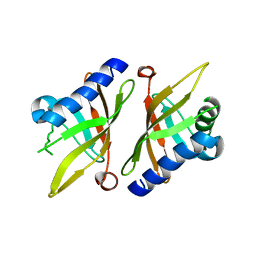 | | Crystal structure of KSI Y57S mutant | | Descriptor: | Ketosteroid Isomerase | | Authors: | Cha, S.S, Oh, B.H, Nam, G.H, Jang, D.S, Lee, T.H, Choi, K.Y. | | Deposit date: | 2001-10-05 | | Release date: | 2002-10-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Maintenance of alpha-helical structures by phenyl rings in the active-site tyrosine triad contributes to catalysis and stability of ketosteroid isomerase from Pseudomonas putida biotype B
Biochemistry, 40, 2001
|
|
3BP3
 
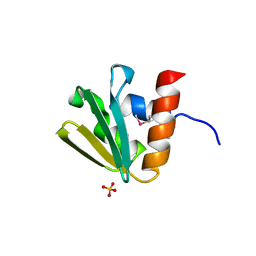 | | Crystal structure of EIIB | | Descriptor: | Glucose-specific phosphotransferase enzyme IIB component, SULFATE ION | | Authors: | Cha, S.S, Jung, H.I, An, Y.J. | | Deposit date: | 2007-12-18 | | Release date: | 2008-11-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Analyses of Mlc-IIBGlc interaction and a plausible molecular mechanism of Mlc inactivation by membrane sequestration.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1ZKJ
 
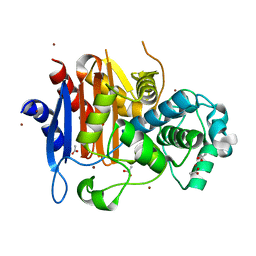 | | Structural Basis for the Extended Substrate Spectrum of CMY-10, a Plasmid-Encoded Class C beta-lactamase | | Descriptor: | ACETIC ACID, ZINC ION, extended-spectrum beta-lactamase | | Authors: | Cha, S.S, Jung, H.I, An, Y.J, Lee, S.H. | | Deposit date: | 2005-05-03 | | Release date: | 2006-04-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the extended substrate spectrum of CMY-10, a plasmid-encoded class C beta-lactamase.
Mol.Microbiol., 60, 2006
|
|
4RMF
 
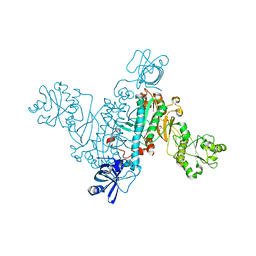 | | Biochemical and structural characterization of mycobacterial aspartyl-tRNA synthetase AspS, a promising TB drug target | | Descriptor: | 2,2-bis(hydroxymethyl)propane-1,3-diol, Aspartate--tRNA(Asp/Asn) ligase, FORMIC ACID | | Authors: | Cox, J.A.G, Gurcha, S.S, Veeraraghavan, U, Besra, G.S, Futterer, K. | | Deposit date: | 2014-10-21 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical and Structural Characterization of Mycobacterial Aspartyl-tRNA Synthetase AspS, a Promising TB Drug Target.
Plos One, 9, 2014
|
|
5X9U
 
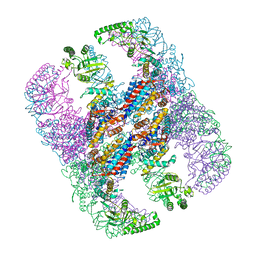 | | Crystal structure of group III chaperonin in the open state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Thermosome, alpha subunit | | Authors: | An, Y.J, Cha, S.S. | | Deposit date: | 2017-03-09 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.001 Å) | | Cite: | Structural and mechanistic characterization of an archaeal-like chaperonin from a thermophilic bacterium
Nat Commun, 8, 2017
|
|
