1GMB
 
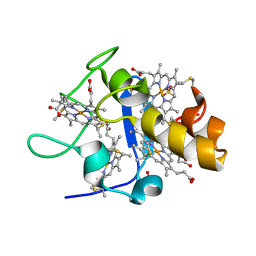 | | Reduced structure of CYTOCHROME C3 FROM DESULFOVIBRIO DESULFURICANS ATCC 27774 at pH 7.6 | | Descriptor: | CYTOCHROME C3, HEME C, SULFATE ION | | Authors: | Bento, I, Louro, R, Matias, P.M, Catarino, T, Baptista, A.M, Soares, C.M, Carrondo, M.A, Turner, D.L, Xavier, A.V. | | Deposit date: | 2001-09-12 | | Release date: | 2002-09-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Component in the Coupled Transfer of Multiple Electrons and Protons in a Monomeric Tetraheme Cytochrome
J.Biol.Chem., 276, 2001
|
|
1GM4
 
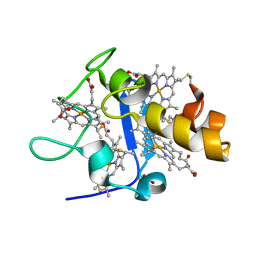 | | OXIDISED STRUCTURE OF CYTOCHROME C3 FROM DESULFOVIBRIO DESULFURICANS ATCC 27774 at pH 7.6 | | Descriptor: | CYTOCHROME C3, HEME C, SULFATE ION | | Authors: | Bento, I, Louro, R, Matias, P.M, Catarino, T, Baptista, A.M, Soares, C.M, Carrondo, M.A, Turner, D.L, Xavier, A.V. | | Deposit date: | 2001-09-10 | | Release date: | 2002-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conformational Component in the Coupled Transfer of Multiple Electrons and Protons in a Monomeric Tetraheme Cytochrome.
J.Biol.Chem., 276, 2001
|
|
1A2I
 
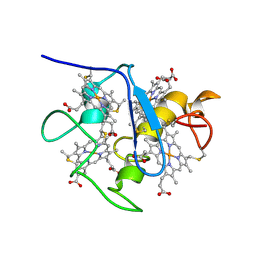 | | SOLUTION STRUCTURE OF DESULFOVIBRIO VULGARIS (HILDENBOROUGH) FERROCYTOCHROME C3, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Messias, A.C, Kastrau, D.H.K, Costa, H.S, Legall, J, Turner, D.L, Santos, H, Xavier, A.V. | | Deposit date: | 1998-01-05 | | Release date: | 1998-07-08 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Desulfovibrio vulgaris (Hildenborough) ferrocytochrome c3: structural basis for functional cooperativity.
J.Mol.Biol., 281, 1998
|
|
7QP5
 
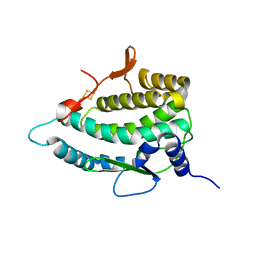 | | Crystal Structure of E. coli FhuF | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferric iron reductase protein FhuF | | Authors: | Trindade, I.B, Rollo, F, Matias, P.M, Moe, E, Louro, R.O. | | Deposit date: | 2022-01-03 | | Release date: | 2023-07-12 | | Last modified: | 2023-07-19 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The structure of a novel ferredoxin: FhuF, a ferric-siderophore reductase from E. coli K-12 with a novel 2Fe-2S cluster coordination
Biorxiv, 2023
|
|
8C4L
 
 | |
6GEH
 
 | | Structure and reactivity of a siderophore-interacting protein from the marine bacterium Shewanella reveals unanticipated functional versatility. | | Descriptor: | DIMETHYL SULFOXIDE, FAD-binding 9, siderophore-interacting domain protein, ... | | Authors: | Trindade, I.B, Silva, J.P.M, Matias, P, Moe, E. | | Deposit date: | 2018-04-26 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure and reactivity of a siderophore-interacting protein from the marine bacteriumShewanellareveals unanticipated functional versatility.
J. Biol. Chem., 294, 2019
|
|
5NA1
 
 | | NADH:quinone oxidoreductase (NDH-II) from Staphylococcus aureus - holoprotein structure - 2.32 A resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MALONATE ION, NADH dehydrogenase-like protein SAOUHSC_00878, ... | | Authors: | Brito, J.A, Athayde, D, Sousa, F.M, Sena, F.V, Pereira, M.M, Archer, M. | | Deposit date: | 2017-02-27 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The key role of glutamate 172 in the mechanism of type II NADH:quinone oxidoreductase of Staphylococcus aureus.
Biochim. Biophys. Acta, 1858, 2017
|
|
5NA4
 
 | | NADH:quinone oxidoreductase (NDH-II) from Staphylococcus aureus - E172S mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH dehydrogenase-like protein SAOUHSC_00878 | | Authors: | Brito, J.A, Athayde, D, Sousa, F.M, Sena, F.V, Pereira, M.M, Archer, M. | | Deposit date: | 2017-02-27 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The key role of glutamate 172 in the mechanism of type II NADH:quinone oxidoreductase of Staphylococcus aureus.
Biochim. Biophys. Acta, 1858, 2017
|
|
3B47
 
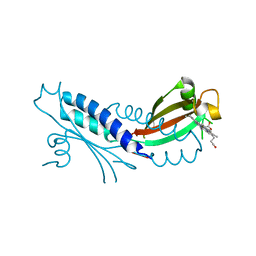 | | Periplasmic sensor domain of chemotaxis protein GSU0582 | | Descriptor: | Methyl-accepting chemotaxis protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pokkuluri, P.R, Schiffer, M. | | Deposit date: | 2007-10-23 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures and solution properties of two novel periplasmic sensor domains with c-type heme from chemotaxis proteins of Geobacter sulfurreducens: implications for signal transduction.
J.Mol.Biol., 377, 2008
|
|
3B42
 
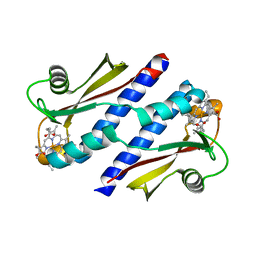 | | Periplasmic sensor domain of chemotaxis protein GSU0935 | | Descriptor: | Methyl-accepting chemotaxis protein, putative, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pokkuluri, P.R, Schiffer, M. | | Deposit date: | 2007-10-23 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures and solution properties of two novel periplasmic sensor domains with c-type heme from chemotaxis proteins of Geobacter sulfurreducens: implications for signal transduction.
J.Mol.Biol., 377, 2008
|
|
1W7O
 
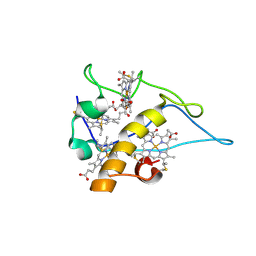 | | cytochrome c3 from Desulfomicrobium baculatus | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Coelho, A.V, Frazao, C, Matias, P.M, Carrondo, M.A. | | Deposit date: | 2004-09-07 | | Release date: | 2004-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Proton-Assisted Two-Electron Transfer in Natural Variants of Tetraheme Cytochromes from Desulfomicrobium Sp
J.Biol.Chem., 279, 2004
|
|
1QN0
 
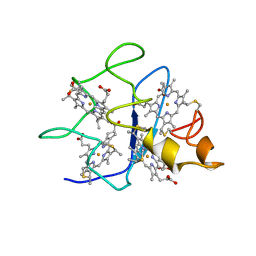 | | SOLUTION STRUCTURE OF DESULFOVIBRIO GIGAS FERROCYTOCHROME C3, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Messias, A.C, Teodoro, M.L, Brennan, L, Legall, J, Santos, H, Xavier, A.V, Turner, D.L. | | Deposit date: | 1999-10-11 | | Release date: | 2000-10-12 | | Last modified: | 2019-11-06 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Network of Functional Cooperativities in Cytochrome C3 from Desulfovibrio Gigas: Solution Structures of the Oxidised and Reduced States
J.Mol.Biol., 298, 2000
|
|
1QN1
 
 | | SOLUTION STRUCTURE OF DESULFOVIBRIO GIGAS FERRICYTOCHROME C3, NMR, 15 STRUCTURES | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Brennan, L, Messias, A.C, Legall, J, Turner, D.L, Xavier, A.V. | | Deposit date: | 1999-10-11 | | Release date: | 2000-10-12 | | Last modified: | 2019-11-06 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Network of Functional Cooperativities in Cytochromes C3 from Desulfovibrio Gigas: Solution Structures of the Oxidised and Reduced States
J.Mol.Biol., 298, 2000
|
|
1UPD
 
 | | Oxidized STRUCTURE OF CYTOCHROME C3 FROM DESULFOVIBRIO DESULFURICANS ATCC 27774 AT PH 7.6 | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Bento, I, Matias, P.M, Baptista, A.M, Da Costa, P.N, Van Dongen, W.M.A.M, Saraiva, L.M, Schneider, T.R, Soares, C.M, Carrondo, M.A. | | Deposit date: | 2003-09-29 | | Release date: | 2004-09-30 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Basis for Redox-Bohr and Cooperative Effects in Cytochrome C3 from Desulfovibrio Desulfuricans Atcc 27774: Crystallographic and Modeling Studies of Oxidized and Reduced High-Resolution Structures at Ph 7.6
Proteins, 54, 2004
|
|
1UP9
 
 | | REDUCED STRUCTURE OF CYTOCHROME C3 FROM DESULFOVIBRIO DESULFURICANS ATCC 27774 AT PH 7.6 | | Descriptor: | CYTOCHROME C3, HEME C, SULFATE ION | | Authors: | Bento, I, Matias, P.M, Baptista, A.M, Da Costa, P.N, Van Dongen, W.M.A.M, Saraiva, L.M, Schneider, T.R, Soares, C.M, Carrondo, M.A. | | Deposit date: | 2003-09-29 | | Release date: | 2004-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Molecular Basis for Redox-Bohr and Cooperative Effects in Cytochrome C3 from Desulfovibrio Desulfuricans Atcc 27774: Crystallographic and Modeling Studies of Oxidized and Reduced High-Resolution Structures at Ph 7.6
Proteins, 54, 2004
|
|
