1ZXQ
 
 | | THE CRYSTAL STRUCTURE OF ICAM-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, INTERCELLULAR ADHESION MOLECULE-2 | | Authors: | Casasnovas, J.M, Springer, T.A, Harrison, S.C, Wang, J.-H. | | Deposit date: | 1997-03-04 | | Release date: | 1997-09-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of ICAM-2 reveals a distinctive integrin recognition surface.
Nature, 387, 1997
|
|
1IC1
 
 | | THE CRYSTAL STRUCTURE FOR THE N-TERMINAL TWO DOMAINS OF ICAM-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, INTERCELLULAR ADHESION MOLECULE-1 | | Authors: | Casasnovas, J.M, Stehle, T, Liu, J.-H, Wang, J.-H, Springer, T.A. | | Deposit date: | 1998-03-09 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A dimeric crystal structure for the N-terminal two domains of intercellular adhesion molecule-1.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
7R4Q
 
 | | The SARS-CoV-2 spike in complex with the 1.29 neutralizing nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Camel-derived nanobody 1.29, ... | | Authors: | Casasnovas, J.M, Melero, R, Arranz, R, Fernandez, L.A. | | Deposit date: | 2022-02-09 | | Release date: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Nanobodies Protecting From Lethal SARS-CoV-2 Infection Target Receptor Binding Epitopes Preserved in Virus Variants Other Than Omicron.
Front Immunol, 13, 2022
|
|
7R4I
 
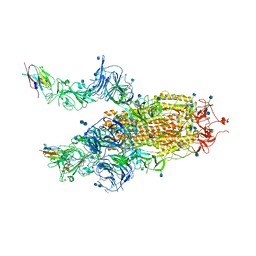 | | The SARS-CoV-2 spike in complex with the 2.15 neutralizing nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Camel-derived nanobody 2.15, ... | | Authors: | Casasnovas, J.M, Melero, R, Arranz, R, Fernandez, L.A. | | Deposit date: | 2022-02-08 | | Release date: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Nanobodies Protecting From Lethal SARS-CoV-2 Infection Target Receptor Binding Epitopes Preserved in Virus Variants Other Than Omicron.
Front Immunol, 13, 2022
|
|
7R4R
 
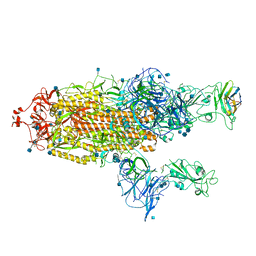 | | The SARS-CoV-2 spike in complex with the 1.10 neutralizing nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Camel-derived nanobody 1.10, ... | | Authors: | Casasnovas, J.M, Melero, R, Arranz, R, Fernandez, L.A. | | Deposit date: | 2022-02-09 | | Release date: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Nanobodies Protecting From Lethal SARS-CoV-2 Infection Target Receptor Binding Epitopes Preserved in Virus Variants Other Than Omicron.
Front Immunol, 13, 2022
|
|
1BQS
 
 | | THE CRYSTAL STRUCTURE OF MUCOSAL ADDRESSIN CELL ADHESION MOLECULE-1 (MADCAM-1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (MUCOSAL ADDRESSIN CELL ADHESION MOLECULE-1) | | Authors: | Tan, K, Casasnovas, J.M, Liu, J.H, Briskin, M.J, Springer, T.A, Wang, J.-H. | | Deposit date: | 1998-08-18 | | Release date: | 1999-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of immunoglobulin superfamily domains 1 and 2 of MAdCAM-1 reveals novel features important for integrin recognition.
Structure, 6, 1998
|
|
4F5C
 
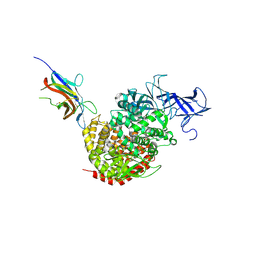 | | Crystal structure of the spike receptor binding domain of a porcine respiratory coronavirus in complex with the pig aminopeptidase N ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Santiago, C, Reguera, J, Gaurav, M, Ordono, D, Enjuanes, L, Casasnovas, J.M. | | Deposit date: | 2012-05-13 | | Release date: | 2012-08-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural bases of coronavirus attachment to host aminopeptidase N and its inhibition by neutralizing antibodies.
Plos Pathog., 8, 2012
|
|
4F2M
 
 | | Crystal structure of a TGEV coronavirus Spike fragment in complex with the TGEV neutralizing monoclonal antibody 1AF10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, ... | | Authors: | Reguera, J, Santiago, C, Mudgal, G, Ordono, D, Enjuanes, L, Casasnovas, J.M. | | Deposit date: | 2012-05-08 | | Release date: | 2012-08-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural bases of coronavirus attachment to host aminopeptidase N and its inhibition by neutralizing antibodies.
Plos Pathog., 8, 2012
|
|
3INB
 
 | | Structure of the measles virus hemagglutinin bound to the CD46 receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin glycoprotein, Membrane cofactor protein, ... | | Authors: | Santiago, C, Celma, M.L, Stehle, T, Casasnovas, J.M. | | Deposit date: | 2009-08-12 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the measles virus hemagglutinin bound to the CD46 receptor
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KAA
 
 | | Structure of Tim-3 in complex with phosphatidylserine | | Descriptor: | 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, CALCIUM ION, Hepatitis A virus cellular receptor 2 | | Authors: | Ballesteros, A, Santiago, C, Casasnovas, J.M. | | Deposit date: | 2009-10-19 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | T cell/transmembrane, Ig, and mucin-3 allelic variants differentially recognize phosphatidylserine and mediate phagocytosis of apoptotic cells.
J.Immunol., 184, 2010
|
|
4OI9
 
 | | Crystal Structure of ICAM-5 D1-D4 ectodomain fragment, Space Group P21 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Recacha, R, Jimenez, D, Tian, L, Barredo, R, Ghamberg, C, Casasnovas, J.M. | | Deposit date: | 2014-01-19 | | Release date: | 2014-07-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of an ICAM-5 ectodomain fragment show electrostatic-based homophilic adhesions.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OIA
 
 | | Crystal Structure of ICAM-5 D1-D4 ectodomain fragment, Space Group P4322 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Recacha, R, Jimenez, D, Tian, L, Barredo, R, Ghamberg, C, Casasnovas, J.M. | | Deposit date: | 2014-01-19 | | Release date: | 2014-07-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structures of an ICAM-5 ectodomain fragment show electrostatic-based homophilic adhesions.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5LDS
 
 | | Structure of the porcine aminopeptidase N ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Santiago, C, Reguera, J, Mudgal, G, Casasnovas, J.M. | | Deposit date: | 2016-06-27 | | Release date: | 2017-04-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric inhibition of aminopeptidase N functions related to tumor growth and virus infection.
Sci Rep, 7, 2017
|
|
5LHD
 
 | | Structure of glycosylated human aminopeptidase N | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Recacha, R, Mudgal, G, Santiago, C, Casasnovas, J.M. | | Deposit date: | 2016-07-11 | | Release date: | 2017-04-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Allosteric inhibition of aminopeptidase N functions related to tumor growth and virus infection.
Sci Rep, 7, 2017
|
|
5LG6
 
 | | Structure of the deglycosylated porcine aminopeptidase N ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ZINC ION | | Authors: | Santiago, C, Reguera, J, Mudgal, G, Casasnovas, J.M. | | Deposit date: | 2016-07-06 | | Release date: | 2017-04-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric inhibition of aminopeptidase N functions related to tumor growth and virus infection.
Sci Rep, 7, 2017
|
|
4OIB
 
 | | Crystal Structure of ICAM-5 D1-D4 ectodomain fragment, Space Group R3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Intercellular adhesion molecule 5, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Recacha, R, Jimenez, D, Tian, L, Barredo, R, Ghamberg, C, Casasnovas, J.M. | | Deposit date: | 2014-01-19 | | Release date: | 2014-07-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Crystal structures of an ICAM-5 ectodomain fragment show electrostatic-based homophilic adhesions.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3BIA
 
 | | Tim-4 in complex with sodium potassium tartrate | | Descriptor: | L(+)-TARTARIC ACID, SODIUM ION, T-cell immunoglobulin and mucin domain-containing protein 4 | | Authors: | Santiago, C, Ballesteros, A, Kaplan, G.G, Freeman, G.J, Casasnovas, J.M. | | Deposit date: | 2007-11-30 | | Release date: | 2008-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of T Cell Immunoglobulin Mucin Protein 4 Show a Metal-Ion-Dependent Ligand Binding Site where Phosphatidylserine Binds.
Immunity, 27, 2007
|
|
3BI9
 
 | | Tim-4 | | Descriptor: | ACETATE ION, T-cell immunoglobulin and mucin domain-containing protein 4 | | Authors: | Santiago, C, Ballesteros, A, Kaplan, G.G, Freeman, G.J, Casasnovas, J.M. | | Deposit date: | 2007-11-30 | | Release date: | 2008-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structures of T Cell Immunoglobulin Mucin Protein 4 Show a Metal-Ion-Dependent Ligand Binding Site where Phosphatidylserine Binds.
Immunity, 27, 2007
|
|
3BIB
 
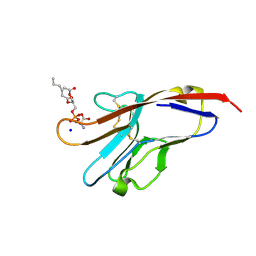 | | Tim-4 in complex with phosphatidylserine | | Descriptor: | 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, SODIUM ION, T-cell immunoglobulin and mucin domain-containing protein 4 | | Authors: | Santiago, C, Ballesteros, A, Kaplan, G.G, Freeman, G.J, Casasnovas, J.M. | | Deposit date: | 2007-11-30 | | Release date: | 2008-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of T Cell Immunoglobulin Mucin Protein 4 Show a Metal-Ion-Dependent Ligand Binding Site where Phosphatidylserine Binds.
Immunity, 27, 2007
|
|
2OR8
 
 | | Tim-1 | | Descriptor: | ACETATE ION, Hepatitis A virus cellular receptor 1 homolog | | Authors: | Santiago, C, Ballesteros, A, Kaplan, G.G, Casasnovas, J.M. | | Deposit date: | 2007-02-02 | | Release date: | 2007-04-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of T Cell Immunoglobulin Mucin Receptors 1 and 2 Reveal Mechanisms for Regulation of Immune Responses by the TIM Receptor Family.
Immunity, 26, 2007
|
|
2OR7
 
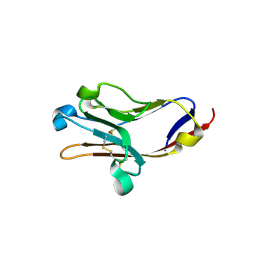 | | Tim-2 | | Descriptor: | ACETATE ION, T-cell immunoglobulin and mucin domain-containing protein 2 | | Authors: | Santiago, C, Ballesteros, A, Kaplan, G.G, Casasnovas, J.M. | | Deposit date: | 2007-02-02 | | Release date: | 2007-04-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of T Cell Immunoglobulin Mucin Receptors 1 and 2 Reveal Mechanisms for Regulation of Immune Responses by the TIM Receptor Family.
Immunity, 26, 2007
|
|
3O74
 
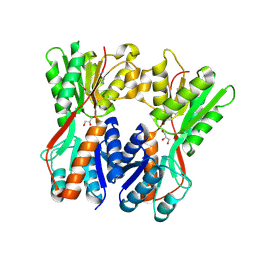 | | Crystal structure of Cra transcriptional dual regulator from Pseudomonas putida | | Descriptor: | Fructose transport system repressor FruR, GLYCEROL | | Authors: | Chavarria, M, Santiago, C, Platero, R, Krell, T, Casasnovas, J.M, de Lorenzo, V. | | Deposit date: | 2010-07-30 | | Release date: | 2011-01-12 | | Last modified: | 2011-12-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fructose 1-phosphate is the preferred effector of the metabolic regulator Cra of Pseudomonas putida
J.Biol.Chem., 286, 2011
|
|
3O75
 
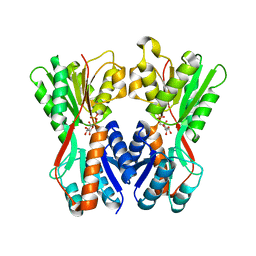 | | Crystal structure of Cra transcriptional dual regulator from Pseudomonas putida in complex with fructose 1-phosphate' | | Descriptor: | 1-O-phosphono-beta-D-fructofuranose, Fructose transport system repressor FruR | | Authors: | Chavarria, M, Santiago, C, Platero, R, Krell, T, Casasnovas, J.M, de Lorenzo, V. | | Deposit date: | 2010-07-30 | | Release date: | 2011-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fructose 1-phosphate is the preferred effector of the metabolic regulator Cra of Pseudomonas putida
J.Biol.Chem., 286, 2011
|
|
3O8E
 
 | | Structure of extracelllar portion of CD46 in complex with Adenovirus type 11 knob | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DITHIANE DIOL, Fiber 36.1 kDa protein, ... | | Authors: | Persson, B.D, Schmitz, N.B, Casasnovas, J.M, Stehle, T. | | Deposit date: | 2010-08-03 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structure of the extracellular portion of CD46 provides insights into its interactions with complement proteins and pathogens.
Plos Pathog., 6, 2010
|
|
1CKL
 
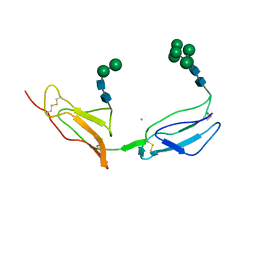 | | N-TERMINAL TWO DOMAINS OF HUMAN CD46 (MEMBRANE COFACTOR PROTEIN, MCP) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Casasnovas, J, Larvie, M, Stehle, T. | | Deposit date: | 1999-04-22 | | Release date: | 1999-06-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of two CD46 domains reveals an extended measles virus-binding surface.
EMBO J., 18, 1999
|
|
