5NGS
 
 | | Crystal structure of human MTH1 in complex with inhibitor 6-[(2-phenylethyl)sulfanyl]-7H-purin-2-amine | | Descriptor: | 6-(2-phenylethylsulfanyl)-7~{H}-purin-2-amine, 7,8-dihydro-8-oxoguanine triphosphatase, ACETATE ION, ... | | Authors: | Gustafsson, R, Rudling, A, Almlof, I, Homan, E, Scobie, M, Warpman Berglund, U, Helleday, T, Carlsson, J, Stenmark, P. | | Deposit date: | 2017-03-20 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fragment-Based Discovery and Optimization of Enzyme Inhibitors by Docking of Commercial Chemical Space.
J. Med. Chem., 60, 2017
|
|
5NGR
 
 | | Crystal structure of human MTH1 in complex with fragment inhibitor 8-(methylsulfanyl)-7H-purin-6-amine | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, 8-methylsulfanyl-7~{H}-purin-6-amine, SULFATE ION | | Authors: | Gustafsson, R, Rudling, A, Almlof, I, Homan, E, Scobie, M, Warpman Berglund, U, Helleday, T, Carlsson, J, Stenmark, P. | | Deposit date: | 2017-03-20 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fragment-Based Discovery and Optimization of Enzyme Inhibitors by Docking of Commercial Chemical Space.
J. Med. Chem., 60, 2017
|
|
5NGT
 
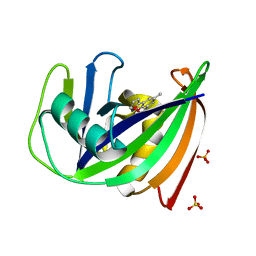 | | Crystal structure of human MTH1 in complex with inhibitor 7-(furan-2-yl)-5-methyl-1,3-benzoxazol-2-amine | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, 7-(furan-2-yl)-5-methyl-1,3-benzoxazol-2-amine, SULFATE ION | | Authors: | Gustafsson, R, Rudling, A, Almlof, I, Homan, E, Scobie, M, Warpman Berglund, U, Helleday, T, Carlsson, J, Stenmark, P. | | Deposit date: | 2017-03-20 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Fragment-Based Discovery and Optimization of Enzyme Inhibitors by Docking of Commercial Chemical Space.
J. Med. Chem., 60, 2017
|
|
7QBB
 
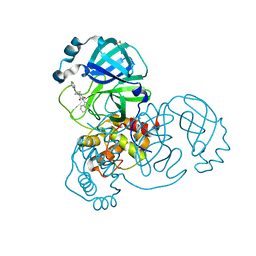 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 18 | | Descriptor: | 3C-like proteinase nsp5, 7-isoquinolin-4-yl-2-phenyl-5,7-diazaspiro[3.4]octane-6,8-dione, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2021-11-18 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7NBT
 
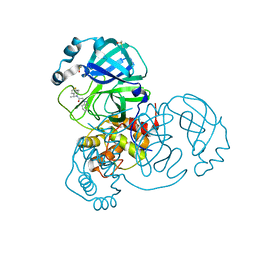 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 21 | | Descriptor: | 2-(benzotriazol-1-yl)-1-[(4~{S})-4-methyl-6,7-dihydro-4~{H}-thieno[3,2-c]pyridin-5-yl]ethanone, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2021-01-27 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7NEO
 
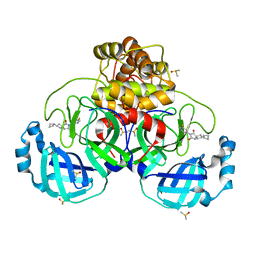 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 15 | | Descriptor: | 2-cyclobutyl-7-(5-fluoropyridin-3-yl)-5,7-diazaspiro[3.4]octane-6,8-dione, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2021-02-04 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7O46
 
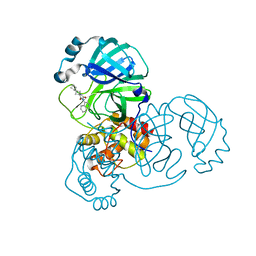 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 17 | | Descriptor: | 2-cyclobutyl-7-isoquinolin-4-yl-5,7-diazaspiro[3.4]octane-6,8-dione, 3C-like proteinase nsp5 | | Authors: | Talibov, V.O. | | Deposit date: | 2021-04-05 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
3NWJ
 
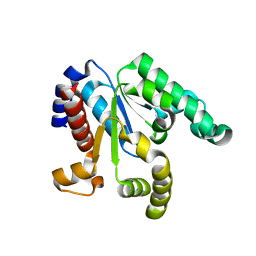 | |
7LD3
 
 | | Cryo-EM structure of the human adenosine A1 receptor-Gi2-protein complex bound to its endogenous agonist and an allosteric ligand | | Descriptor: | ADENOSINE, Chimera protein of Muscarinic acetylcholine receptor M4 and Adenosine receptor A1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Draper-Joyce, C.J, Danev, R, Thal, D.M, Christopoulos, A, Glukhova, A. | | Deposit date: | 2021-01-12 | | Release date: | 2021-09-08 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Positive allosteric mechanisms of adenosine A 1 receptor-mediated analgesia.
Nature, 597, 2021
|
|
7LD4
 
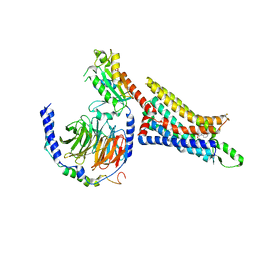 | | Cryo-EM structure of the human adenosine A1 receptor-Gi2-protein complex bound to its endogenous agonist | | Descriptor: | ADENOSINE, Chimera protein of Muscarinic acetylcholine receptor M4 and Adenosine receptor A1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Draper-Joyce, C.J, Danev, R, Thal, D.M, Christopoulos, A, Glukhova, A. | | Deposit date: | 2021-01-12 | | Release date: | 2021-09-08 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Positive allosteric mechanisms of adenosine A 1 receptor-mediated analgesia.
Nature, 597, 2021
|
|
7AU4
 
 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 3 | | Descriptor: | (3~{S})-6-chloranyl-3'-(1,2-oxazol-3-ylmethyl)spiro[1,2-dihydroindene-3,5'-imidazolidine]-2',4'-dione, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2020-11-02 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7B5Z
 
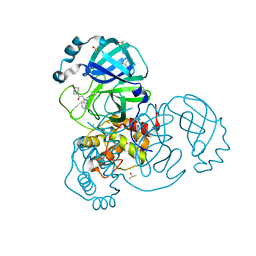 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 6 | | Descriptor: | 2-(1H-benzo[d][1,2,3]triazol-1-yl)-1-(4-methylenepiperidin-1-yl)ethan-1-one, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2020-12-07 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7B2U
 
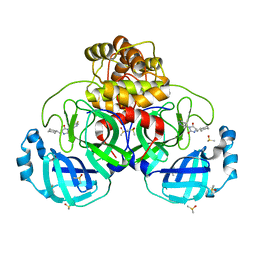 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 1 | | Descriptor: | (5S)-5-(cyclohexylmethyl)-3-(5-fluoropyridin-3-yl)imidazolidine-2,4-dione, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2020-11-27 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7B77
 
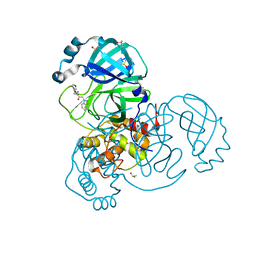 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 8 | | Descriptor: | 2-(benzotriazol-1-yl)-~{N}-ethyl-~{N}-(furan-3-ylmethyl)ethanamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2020-12-09 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7B2J
 
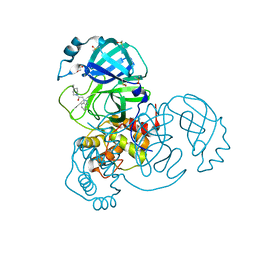 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 5 | | Descriptor: | 2-(1H-1,2,3-benzotriazol-1-yl)-1-(4-methylpiperidin-1-yl)ethan-1-one, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Talibov, V.O. | | Deposit date: | 2020-11-27 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7BIJ
 
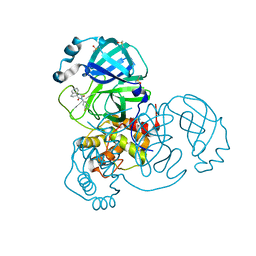 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 13 | | Descriptor: | (3~{S})-3'-(5-fluoranylpyridin-3-yl)spiro[1,2-dihydroindene-3,5'-imidazolidine]-2',4'-dione, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2021-01-12 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7OIL
 
 | | mPI3Kd in complex with compound 58 | | Descriptor: | 2-[(1S)-1-cyclopropylethyl]-5-[4-methyl-2-[[6-(2-oxidanylidenepyrrolidin-1-yl)pyridin-2-yl]amino]-1,3-thiazol-5-yl]-7-methylsulfonyl-3H-isoindol-1-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, SODIUM ION | | Authors: | Petersen, J. | | Deposit date: | 2021-05-11 | | Release date: | 2021-07-14 | | Last modified: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of AZD8154, a Dual PI3K gamma delta Inhibitor for the Treatment of Asthma.
J.Med.Chem., 64, 2021
|
|
7OI4
 
 | | mPI3Kd in complex with compound 12 | | Descriptor: | N-[5-[2-[(1S)-1-cyclopropylethyl]-7-[[4-[(dimethylamino)methyl]phenyl]sulfamoyl]-1-oxidanylidene-3H-isoindol-5-yl]-4-methyl-1,3-thiazol-2-yl]ethanamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Petersen, J. | | Deposit date: | 2021-05-11 | | Release date: | 2021-07-14 | | Last modified: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of AZD8154, a Dual PI3K gamma delta Inhibitor for the Treatment of Asthma.
J.Med.Chem., 64, 2021
|
|
7OIJ
 
 | | mPI3Kd in complex with an inhibitor | | Descriptor: | 6-[[5-[2-[(1S)-1-cyclopropylethyl]-7-(methylsulfamoyl)-1-oxidanylidene-3H-isoindol-5-yl]-4-methyl-1,3-thiazol-2-yl]amino]-N-[3-(dimethylamino)propyl]pyridine-2-carboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, SODIUM ION, ... | | Authors: | Petersen, J. | | Deposit date: | 2021-05-11 | | Release date: | 2021-07-14 | | Last modified: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of AZD8154, a Dual PI3K gamma delta Inhibitor for the Treatment of Asthma.
J.Med.Chem., 64, 2021
|
|
7OIS
 
 | | mPI3Kd in complex with compound 7 | | Descriptor: | N-[5-[2-[(1S)-1-cyclopropylethyl]-7-[(3-methylsulfonylphenyl)sulfamoyl]-1-oxidanylidene-3H-isoindol-5-yl]-4-methyl-1,3-thiazol-2-yl]ethanamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, SODIUM ION | | Authors: | Petersen, J. | | Deposit date: | 2021-05-12 | | Release date: | 2021-07-14 | | Last modified: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of AZD8154, a Dual PI3K gamma delta Inhibitor for the Treatment of Asthma.
J.Med.Chem., 64, 2021
|
|
7Q6Q
 
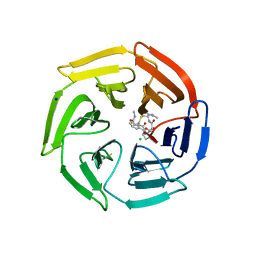 | | Keap1 compound complex | | Descriptor: | (5S,8R)-N,N-dimethyl-8-[[(2S)-1-[4-(methylamino)-4-oxidanylidene-butanoyl]pyrrolidin-2-yl]carbonylamino]-7,11-bis(oxidanylidene)-10-oxa-3-thia-6-azabicyclo[10.4.0]hexadeca-1(12),13,15-triene-5-carboxamide, CHLORIDE ION, Kelch-like ECH-associated protein 1, ... | | Authors: | Johansson, P. | | Deposit date: | 2021-11-09 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Importance of Binding Site Hydration and Flexibility Revealed When Optimizing a Macrocyclic Inhibitor of the Keap1-Nrf2 Protein-Protein Interaction.
J.Med.Chem., 65, 2022
|
|
7Q5H
 
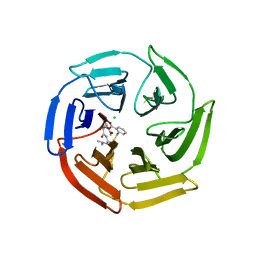 | | Keap1 compound complex | | Descriptor: | (3S,5S,8R)-8-[[(2S)-1-ethanoylpyrrolidin-2-yl]carbonylamino]-N,N-dimethyl-3,7,11-tris(oxidanylidene)-10-oxa-3$l^{4}-thia-6-azabicyclo[10.4.0]hexadeca-1(16),12,14-triene-5-carboxamide, CHLORIDE ION, Kelch-like ECH-associated protein 1 | | Authors: | Johansson, P. | | Deposit date: | 2021-11-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Importance of Binding Site Hydration and Flexibility Revealed When Optimizing a Macrocyclic Inhibitor of the Keap1-Nrf2 Protein-Protein Interaction.
J.Med.Chem., 65, 2022
|
|
7Q96
 
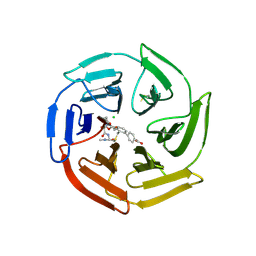 | | Keap1 compound complex | | Descriptor: | 4-[(5S,8R)-5-(dimethylcarbamoyl)-8-[[(2S)-1-ethanoylpyrrolidin-2-yl]carbonylamino]-7,11-bis(oxidanylidene)-10-oxa-3-thia-6-azabicyclo[10.4.0]hexadeca-1(16),12,14-trien-16-yl]benzoic acid, CHLORIDE ION, Kelch-like ECH-associated protein 1 | | Authors: | Johansson, P. | | Deposit date: | 2021-11-12 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.415 Å) | | Cite: | Importance of Binding Site Hydration and Flexibility Revealed When Optimizing a Macrocyclic Inhibitor of the Keap1-Nrf2 Protein-Protein Interaction.
J.Med.Chem., 65, 2022
|
|
7Q6S
 
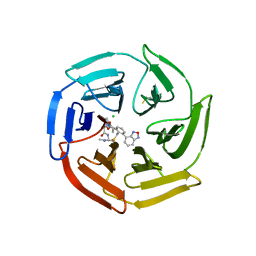 | | Keap1 compound complex | | Descriptor: | (5S,8R)-16-(2,1,3-benzoxadiazol-4-yl)-8-[[(2S)-1-ethanoylpyrrolidin-2-yl]carbonylamino]-N,N-dimethyl-7,11-bis(oxidanylidene)-10-oxa-3-thia-6-azabicyclo[10.4.0]hexadeca-1(16),12,14-triene-5-carboxamide, CHLORIDE ION, Kelch-like ECH-associated protein 1 | | Authors: | Johansson, P. | | Deposit date: | 2021-11-09 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.143 Å) | | Cite: | Importance of Binding Site Hydration and Flexibility Revealed When Optimizing a Macrocyclic Inhibitor of the Keap1-Nrf2 Protein-Protein Interaction.
J.Med.Chem., 65, 2022
|
|
7Q8R
 
 | | Keap1 compound complex | | Descriptor: | 3-[(5S,8R)-5-(dimethylcarbamoyl)-8-[[(2S)-1-ethanoylpyrrolidin-2-yl]carbonylamino]-7,11-bis(oxidanylidene)-10-oxa-3-thia-6-azabicyclo[10.4.0]hexadeca-1(16),12,14-trien-16-yl]benzoic acid, CHLORIDE ION, Kelch-like ECH-associated protein 1 | | Authors: | Johansson, P. | | Deposit date: | 2021-11-11 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | Importance of Binding Site Hydration and Flexibility Revealed When Optimizing a Macrocyclic Inhibitor of the Keap1-Nrf2 Protein-Protein Interaction.
J.Med.Chem., 65, 2022
|
|
