3IJT
 
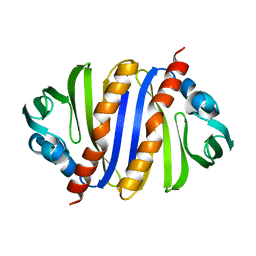 | | Structural Characterization of SMU.440, a Hypothetical Protein from Streptococcus mutans | | Descriptor: | Putative uncharacterized protein | | Authors: | Nan, J, Brostromer, E, Kristensen, O, Su, X.-D. | | Deposit date: | 2009-08-05 | | Release date: | 2009-08-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.377 Å) | | Cite: | Bioinformatics and structural characterization of a hypothetical protein from Streptococcus mutans: implication of antibiotic resistance
Plos One, 4, 2009
|
|
2BB0
 
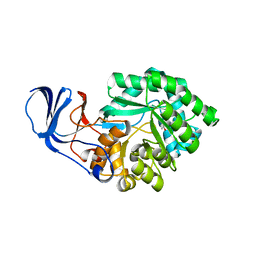 | |
2G3F
 
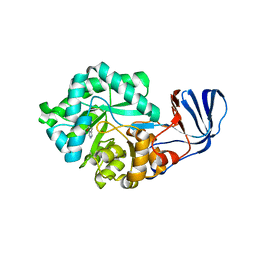 | | Crystal Structure of imidazolonepropionase complexed with imidazole-4-acetic acid sodium salt, a substrate homologue | | Descriptor: | 2H-IMIDAZOL-4-YLACETIC ACID, Imidazolonepropionase, ZINC ION | | Authors: | Yu, Y, Liang, Y.H, Su, X.D. | | Deposit date: | 2006-02-19 | | Release date: | 2006-09-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A catalytic mechanism revealed by the crystal structures of the imidazolonepropionase from Bacillus subtilis
J.Biol.Chem., 281, 2006
|
|
3FYS
 
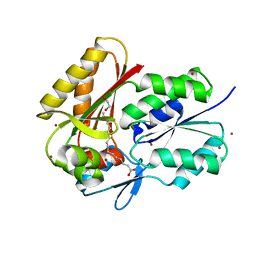 | | Crystal Structure of DegV, a fatty acid binding protein from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, PALMITIC ACID, ... | | Authors: | Nan, J, Zhou, Y.F, Yang, C. | | Deposit date: | 2009-01-23 | | Release date: | 2009-05-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a fatty acid-binding protein from Bacillus subtilis determined by sulfur-SAD phasing using in-house chromium radiation
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3CP7
 
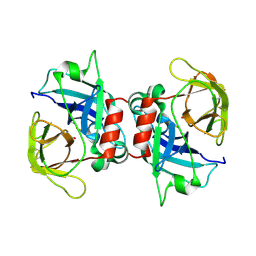 | |
3EXS
 
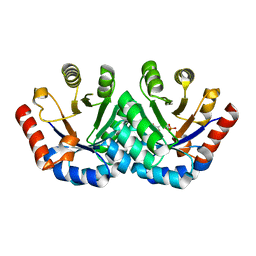 | | Crystal structure of KGPDC from Streptococcus mutans in complex with D-R5P | | Descriptor: | RIBULOSE-5-PHOSPHATE, RmpD (Hexulose-6-phosphate synthase) | | Authors: | Li, G.L, Liu, X, Wang, K.T, Li, L.F, Su, X.D. | | Deposit date: | 2008-10-17 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Open-closed conformational change revealed by the crystal structures of 3-keto-L-gulonate 6-phosphate decarboxylase from Streptococcus mutans
Biochem.Biophys.Res.Commun., 381, 2009
|
|
3EXT
 
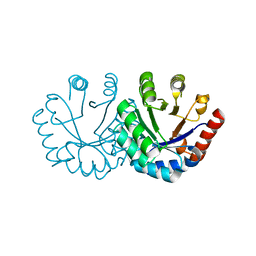 | | Crystal structure of KGPDC from Streptococcus mutans | | Descriptor: | MAGNESIUM ION, RmpD (Hexulose-6-phosphate synthase) | | Authors: | Liu, X, Li, G.L, Li, L.F, Su, X.D. | | Deposit date: | 2008-10-17 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Open-closed conformational change revealed by the crystal structures of 3-keto-L-gulonate 6-phosphate decarboxylase from Streptococcus mutans
Biochem.Biophys.Res.Commun., 381, 2009
|
|
3EXR
 
 | | Crystal structure of KGPDC from Streptococcus mutans | | Descriptor: | RmpD (Hexulose-6-phosphate synthase) | | Authors: | Li, G.L, Liu, X, Li, L.F, Su, X.D. | | Deposit date: | 2008-10-16 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Open-closed conformational change revealed by the crystal structures of 3-keto-L-gulonate 6-phosphate decarboxylase from Streptococcus mutans
Biochem.Biophys.Res.Commun., 381, 2009
|
|
4E2A
 
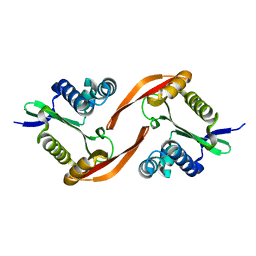 | |
4ETI
 
 | |
