3DA4
 
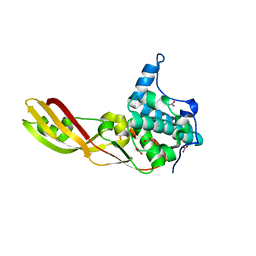 | | Crystal Structure of Colicin M, a Novel Phosphatase Specifically Imported by Escherichia Coli | | Descriptor: | Colicin-M, NITRATE ION | | Authors: | Zeth, K, Albrecht, R, Romer, C, Braun, V. | | Deposit date: | 2008-05-28 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of colicin M, a novel phosphatase specifically imported by Escherichia coli
J.Biol.Chem., 283, 2008
|
|
3DA3
 
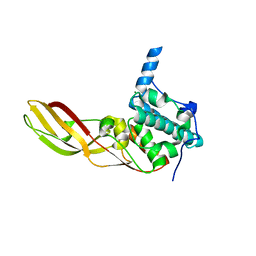 | | Crystal Structure of Colicin M, A Novel Phosphatase Specifically Imported by Escherichia Coli | | Descriptor: | Colicin-M, MAGNESIUM ION | | Authors: | Zeth, K, Albrecht, R, Romer, C, Braun, V. | | Deposit date: | 2008-05-28 | | Release date: | 2008-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of colicin M, a novel phosphatase specifically imported by Escherichia coli
J.Biol.Chem., 283, 2008
|
|
1FI1
 
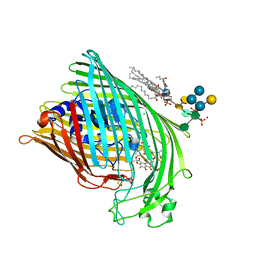 | | FhuA in complex with lipopolysaccharide and rifamycin CGP4832 | | Descriptor: | 3-HYDROXY-TETRADECANOIC ACID, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, DIPHOSPHATE, ... | | Authors: | Ferguson, A.D, Koedding, J, Boes, C, Walker, G, Coulton, J.W, Diederichs, K, Braun, V, Welte, W. | | Deposit date: | 2000-08-03 | | Release date: | 2001-08-29 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Active transport of an antibiotic rifamycin derivative by the outer-membrane protein FhuA.
Structure, 9, 2001
|
|
2XMX
 
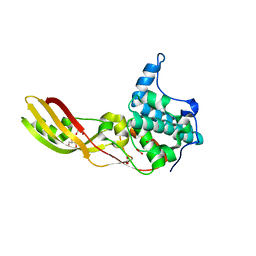 | | High resolution structure of Colicin M | | Descriptor: | CARBONATE ION, COLICIN-M, GLYCEROL | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Braun, V. | | Deposit date: | 2010-07-29 | | Release date: | 2010-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Activation of Colicin M by the Fkpa Prolyl Cis- Trans Isomerase/Chaperone.
J.Biol.Chem., 286, 2011
|
|
2XTR
 
 | | Structure of the P176A Colicin M mutant from E. coli | | Descriptor: | COLICIN-M, NITRATE ION | | Authors: | Helbig, S, Patzer, S.I, Braun, V, Zeth, K. | | Deposit date: | 2010-10-12 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Activation of Colicin M by the Fkpa Prolyl Cis- Trans Isomerase/Chaperone.
J.Biol.Chem., 286, 2011
|
|
2XTQ
 
 | |
4AEQ
 
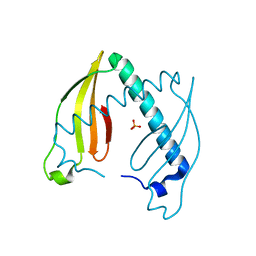 | | Crystal structure of the dimeric immunity protein Cmi solved by direct methods (Arcimboldo) | | Descriptor: | COLICIN-M IMMUNITY PROTEIN, PHOSPHATE ION | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Uson, I, Braun, V. | | Deposit date: | 2012-01-12 | | Release date: | 2012-01-25 | | Last modified: | 2012-04-18 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | The Crystal Structure of the Dimeric Colicin M Immunity
J.Struct.Biol., 178, 2012
|
|
4ARJ
 
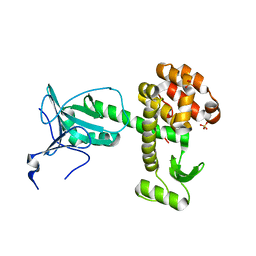 | | Crystal structure of a pesticin (translocation and receptor binding domain) from Y. pestis and T4-lysozyme chimera | | Descriptor: | PESTICIN, LYSOZYME, SULFATE ION | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Braun, V. | | Deposit date: | 2012-04-24 | | Release date: | 2012-05-09 | | Last modified: | 2017-03-15 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | Structure and Mechanistic Studies of Pesticin, a Bacterial Homolog of Phage Lysozymes.
J.Biol.Chem., 287, 2012
|
|
4ARL
 
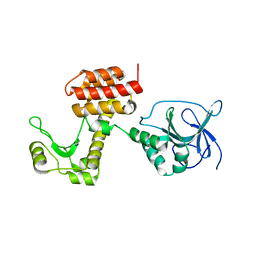 | | Structure of the inactive pesticin D207A mutant | | Descriptor: | PESTICIN | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Braun, V. | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-16 | | Last modified: | 2012-09-05 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structure and Mechanistic Studies of Pesticin, a Bacterial Homolog of Phage Lysozymes.
J.Biol.Chem., 287, 2012
|
|
4ARM
 
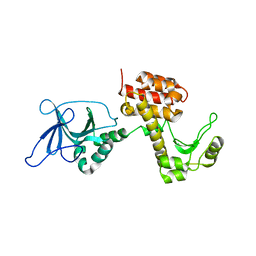 | | Structure of the inactive pesticin T201A mutant | | Descriptor: | PESTICIN | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Braun, V. | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-02 | | Last modified: | 2012-07-18 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structure and Mechanistic Studies of Pesticin, a Bacterial Homolog of Phage Lysozymes.
J.Biol.Chem., 287, 2012
|
|
4ARP
 
 | | Structure of the inactive pesticin E178A mutant | | Descriptor: | PESTICIN | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Braun, V. | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.296 Å) | | Cite: | Structure and Mechanistic Studies of Pesticin, a Bacterial Homolog of Phage Lysozymes.
J.Biol.Chem., 287, 2012
|
|
4ARQ
 
 | | Structure of the pesticin S89C, S285C double mutant | | Descriptor: | MAGNESIUM ION, PESTICIN | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Braun, V. | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Mechanistic Studies of Pesticin, a Bacterial Homolog of Phage Lysozymes.
J.Biol.Chem., 287, 2012
|
|
1ESZ
 
 | | STRUCTURE OF THE PERIPLASMIC FERRIC SIDEROPHORE BINDING PROTEIN FHUD COMPLEXED WITH COPROGEN | | Descriptor: | COPROGEN, FERRICHROME-BINDING PERIPLASMIC PROTEIN | | Authors: | Clarke, T.E, Braun, V, Winkelmann, G, Tari, L.W, Vogel, H.J. | | Deposit date: | 2000-04-11 | | Release date: | 2002-04-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystallographic structures of the Escherichia coli periplasmic protein FhuD bound to hydroxamate-type siderophores and the antibiotic albomycin.
J.Biol.Chem., 277, 2002
|
|
1K2V
 
 | | E. COLI PERIPLASMIC PROTEIN FHUD COMPLEXED WITH DESFERAL | | Descriptor: | DEFEROXAMINE MESYLATE FE(III) COMPLEX, Ferrichrome-binding periplasmic protein | | Authors: | Clarke, T.E, Braun, V, Winkelmann, G, Tari, L.W, Vogel, H.J. | | Deposit date: | 2001-09-29 | | Release date: | 2002-04-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | X-ray crystallographic structures of the Escherichia coli periplasmic protein FhuD bound to hydroxamate-type siderophores and the antibiotic albomycin.
J.Biol.Chem., 277, 2002
|
|
1K7S
 
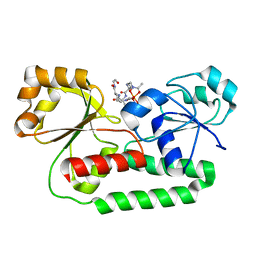 | | FhuD complexed with albomycin-delta 2 | | Descriptor: | DELTA-2-ALBOMYCIN A1, Ferrichrome-binding periplasmic protein | | Authors: | Clarke, T.E, Braun, V, Winkelmann, G, Tari, L.W, Vogel, H.J. | | Deposit date: | 2001-10-21 | | Release date: | 2002-04-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray crystallographic structures of the Escherichia coli periplasmic protein FhuD bound to hydroxamate-type siderophores and the antibiotic albomycin.
J.Biol.Chem., 277, 2002
|
|
1QJQ
 
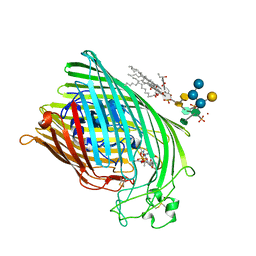 | | FERRIC HYDROXAMATE RECEPTOR FROM ESCHERICHIA COLI (FHUA) | | Descriptor: | 3-HYDROXY-TETRADECANOIC ACID, DIPHOSPHATE, FERRIC HYDROXAMATE RECEPTOR, ... | | Authors: | Ferguson, A.D, Braun, V, Fiedler, H.-P, Coulton, J.W, Diederichs, K, Welte, W. | | Deposit date: | 1999-06-29 | | Release date: | 2000-06-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of the antibiotic albomycin in complex with the outer membrane transporter FhuA.
Protein Sci., 9, 2000
|
|
1QKC
 
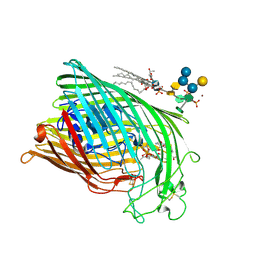 | | ESCHERICHIA COLI FERRIC HYDROXAMATE UPTAKE RECEPTOR (FHUA) IN COMPLEX DELTA TWO-ALBOMYCIN | | Descriptor: | 3-HYDROXY-TETRADECANOIC ACID, DELTA-2-ALBOMYCIN A1, DIPHOSPHATE, ... | | Authors: | Ferguson, A.D, Braun, V, Fiedler, H.-P, Coulton, J.W, Diederichs, K, Welte, W. | | Deposit date: | 1999-07-18 | | Release date: | 2000-06-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the antibiotic albomycin in complex with the outer membrane transporter FhuA.
Protein Sci., 9, 2000
|
|
5CPC
 
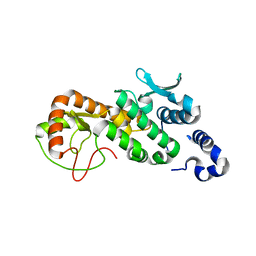 | |
5CQ9
 
 | |
4AQN
 
 | | Crystal structure of pesticin from Y. pestis | | Descriptor: | MAGNESIUM ION, PESTICIN | | Authors: | Zeth, K, Albrecht, R. | | Deposit date: | 2012-04-19 | | Release date: | 2012-05-02 | | Last modified: | 2012-07-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure and Mechanistic Studies of Pesticin, a Bacterial Homolog of Phage Lysozymes.
J.Biol.Chem., 287, 2012
|
|
4C46
 
 | | ANDREI-N-LVPAS fused to GCN4 adaptors | | Descriptor: | BROMIDE ION, GENERAL CONTROL PROTEIN GCN4 | | Authors: | Albrecht, R, Alva, V, Ammelburg, M, Baer, K, Basina, E, Boichenko, I, Bonhoeffer, F, Braun, V, Chaubey, M, Chauhan, N, Chellamuthu, V.R, Coles, M, Deiss, S, Ewers, C.P, Forouzan, D, Fuchs, A, Groemping, Y, Hartmann, M.D, Hernandez Alvarez, B, Jeganantham, A, Kalev, I, Koenninger, U, Koiwai, K, Kopec, K.O, Korycinski, M, Laudenbach, B, Lehmann, K, Leo, J.C, Linke, D, Marialke, J, Martin, J, Mechelke, M, Michalik, M, Noll, A, Patzer, S.I, Scharfenberg, F, Schueckel, M, Shahid, S.A, Sulz, E, Ursinus, A, Wuertenberger, S, Zhu, H. | | Deposit date: | 2013-08-30 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Your Personalized Protein Structure: Andrei N. Lupas Fused to GCN4 Adaptors.
J.Struct.Biol., 186, 2014
|
|
