5A9Q
 
 | | Human nuclear pore complex | | Descriptor: | NUCLEAR PORE COMPLEX PROTEIN NUP107, NUCLEAR PORE COMPLEX PROTEIN NUP133, NUCLEAR PORE COMPLEX PROTEIN NUP155, ... | | Authors: | von Appen, A, Kosinski, J, Sparks, L, Ori, A, DiGuilio, A, Vollmer, B, Mackmull, M, Banterle, N, Parca, L, Kastritis, P, Buczak, K, Mosalaganti, S, Hagen, W, Andres-Pons, A, Lemke, E.A, Bork, P, Antonin, W, Glavy, J.S, Bui, K.H, Beck, M. | | Deposit date: | 2015-07-22 | | Release date: | 2015-09-30 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | In Situ Structural Analysis of the Human Nuclear Pore Complex
Nature, 526, 2015
|
|
1D8B
 
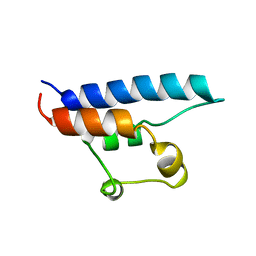 | | NMR STRUCTURE OF THE HRDC DOMAIN FROM SACCHAROMYCES CEREVISIAE RECQ HELICASE | | Descriptor: | SGS1 RECQ HELICASE | | Authors: | Liu, Z, Macias, M.J, Bottomley, M.J, Stier, G, Linge, J.P, Nilges, M, Bork, P, Sattler, M. | | Deposit date: | 1999-10-21 | | Release date: | 2000-01-10 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the HRDC domain and implications for the Werner and Bloom syndrome proteins.
Structure Fold.Des., 7, 1999
|
|
1SGG
 
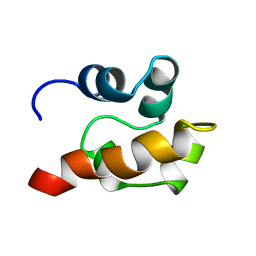 | | THE SOLUTION STRUCTURE OF SAM DOMAIN FROM THE RECEPTOR TYROSINE KINASE EPHB2, NMR, 10 STRUCTURES | | Descriptor: | EPHRIN TYPE-B RECEPTOR 2 | | Authors: | Smalla, M, Schmieder, P, Kelly, M, Ter Laak, A, Krause, G, Ball, L, Wahl, M, Bork, P, Oschkinat, H. | | Deposit date: | 1999-01-08 | | Release date: | 1999-10-06 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the receptor tyrosine kinase EphB2 SAM domain and identification of two distinct homotypic interaction sites.
Protein Sci., 8, 1999
|
|
7OOC
 
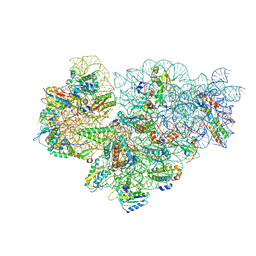 | | Mycoplasma pneumoniae 30S subunit of ribosomes in chloramphenicol-treated cells | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Xue, L, Lenz, S, Rappsilber, J, Mahamid, J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-05-25 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Visualizing translation dynamics at atomic detail inside a bacterial cell.
Nature, 610, 2022
|
|
7OOD
 
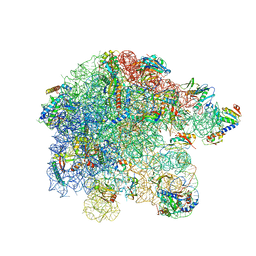 | | Mycoplasma pneumoniae 50S subunit of ribosomes in chloramphenicol-treated cells | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Xue, L, Lenz, S, Rappsilber, J, Mahamid, J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-05-25 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Visualizing translation dynamics at atomic detail inside a bacterial cell.
Nature, 610, 2022
|
|
5FM7
 
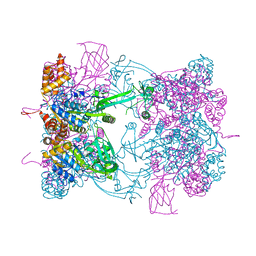 | | Double-heterohexameric rings of full-length Rvb1(ADP)Rvb2(ADP) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RVB1, RVB2 | | Authors: | Silva-Martin, N, Dauden, M.I, Glatt, S, Hoffmann, N.A, Mueller, C.W. | | Deposit date: | 2015-11-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | The Combination of X-Ray Crystallography and Cryo-Electron Microscopy Provides Insight Into the Overall Architecture of the Dodecameric Rvb1/Rvb2 Complex.
Plos One, 11, 2016
|
|
5FM6
 
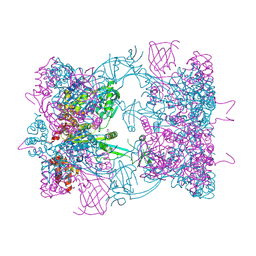 | | Double-heterohexameric rings of full-length Rvb1(ADP)Rvb2(apo) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, RVB1, ... | | Authors: | Silva-Martin, N, Dauden, M.I, Glatt, S, Hoffmann, N.A, Mueller, C.W. | | Deposit date: | 2015-11-02 | | Release date: | 2016-01-20 | | Method: | X-RAY DIFFRACTION (2.997 Å) | | Cite: | The Combination of X-Ray Crystallography and Cryo-Electron Microscopy Provides Insight Into the Overall Architecture of the Dodecameric Rvb1/Rvb2 Complex.
Plos One, 11, 2016
|
|
4IPA
 
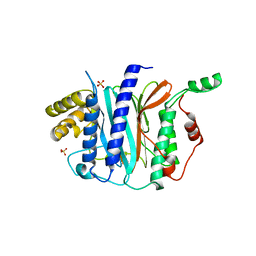 | | Structure of a thermophilic Arx1 | | Descriptor: | Putative curved DNA-binding protein, SULFATE ION | | Authors: | Bange, G, Sinning, I. | | Deposit date: | 2013-01-09 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Consistent mutational paths predict eukaryotic thermostability.
BMC Evol Biol, 13, 2013
|
|
3NSU
 
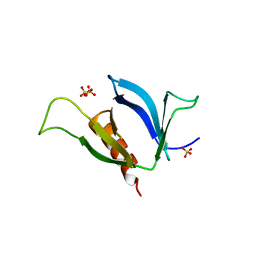 | | A Systematic Screen for Protein-Lipid Interactions in Saccharomyces cerevisiae | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate-binding protein SLM1, SULFATE ION | | Authors: | Gallego, O, Fernandez-Tornero, C, Aguilar-Gurrieri, C, Muller, C, Gavin, A.C. | | Deposit date: | 2010-07-02 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A systematic screen for protein-lipid interactions in Saccharomyces cerevisiae.
Mol. Syst. Biol., 6, 2010
|
|
7PAI
 
 | | 70S ribosome with P-site tRNA in Mycoplasma pneumoniae cells | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Xue, L, Lenz, S, Rappsilber, J, Mahamid, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-05-25 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Visualizing translation dynamics at atomic detail inside a bacterial cell.
Nature, 610, 2022
|
|
7PAK
 
 | | 70S ribosome with EF-Tu-tRNA and P-site tRNA in Mycoplasma pneumoniae cells | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Xue, L, Lenz, S, Rappsilber, J, Mahamid, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-05-25 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Visualizing translation dynamics at atomic detail inside a bacterial cell.
Nature, 610, 2022
|
|
7PAH
 
 | | 70S ribosome with P- and E-site tRNAs in Mycoplasma pneumoniae cells | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Xue, L, Lenz, S, Rappsilber, J, Mahamid, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-05-25 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Visualizing translation dynamics at atomic detail inside a bacterial cell.
Nature, 610, 2022
|
|
7PAU
 
 | | free 50S in complex with ribosome recycling factor in untreated Mycoplasma pneumoniae cells | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Xue, L, Lenz, S, Rappsilber, J, Mahamid, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-05-25 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | Visualizing translation dynamics at atomic detail inside a bacterial cell.
Nature, 610, 2022
|
|
7PAM
 
 | | 70S ribosome with A*- and P/E-site tRNAs in Mycoplasma pneumoniae cells | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Xue, L, Lenz, S, Rappsilber, J, Mahamid, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-05-25 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Visualizing translation dynamics at atomic detail inside a bacterial cell.
Nature, 610, 2022
|
|
7PIA
 
 | | 70S ribosome with A/P- and P/E-site tRNAs in spectinomycin-treated Mycoplasma pneumoniae cells | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Xue, L, Lenz, S, Rappsilber, J, Mahamid, J. | | Deposit date: | 2021-08-19 | | Release date: | 2022-05-25 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (13.6 Å) | | Cite: | Visualizing translation dynamics at atomic detail inside a bacterial cell.
Nature, 610, 2022
|
|
7PHA
 
 | | 70S ribosome with EF-Tu-tRNA and P-site tRNA in chloramphenicol-treated Mycoplasma pneumoniae cells | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Xue, L, Lenz, S, Rappsilber, J, Mahamid, J. | | Deposit date: | 2021-08-16 | | Release date: | 2022-05-25 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Visualizing translation dynamics at atomic detail inside a bacterial cell.
Nature, 610, 2022
|
|
7PAS
 
 | | 70S ribosome with P/E-site tRNA in Mycoplasma pneumoniae cells | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Xue, L, Lenz, S, Rappsilber, J, Mahamid, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-05-25 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Visualizing translation dynamics at atomic detail inside a bacterial cell.
Nature, 610, 2022
|
|
7PI9
 
 | | 70S ribosome with EF-Tu-tRNA and P-site tRNA in spectinomycin-treated Mycoplasma pneumoniae cells | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Xue, L, Lenz, S, Rappsilber, J, Mahamid, J. | | Deposit date: | 2021-08-19 | | Release date: | 2022-05-25 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Visualizing translation dynamics at atomic detail inside a bacterial cell.
Nature, 610, 2022
|
|
7P6Z
 
 | | Mycoplasma pneumoniae 70S ribosome in untreated cells | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Xue, L, Lenz, S, Rappsilber, J, Mahamid, J. | | Deposit date: | 2021-07-18 | | Release date: | 2022-05-25 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Visualizing translation dynamics at atomic detail inside a bacterial cell.
Nature, 610, 2022
|
|
7PAQ
 
 | | 70S ribosome with EF-G, A/P- and P/E-site tRNAs in Mycoplasma pneumoniae cells | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Xue, L, Lenz, S, Rappsilber, J, Mahamid, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-05-25 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Visualizing translation dynamics at atomic detail inside a bacterial cell.
Nature, 610, 2022
|
|
7PIQ
 
 | | 70S ribosome with A- and P-site tRNAs in pseudouridimycin-treated Mycoplasma pneumoniae cells | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Xue, L, Lenz, S, Rappsilber, J, Mahamid, J. | | Deposit date: | 2021-08-23 | | Release date: | 2022-05-25 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (9.7 Å) | | Cite: | Visualizing translation dynamics at atomic detail inside a bacterial cell.
Nature, 610, 2022
|
|
7PAR
 
 | | 70S ribosome with EF-G, ap/P- and pe/E-site tRNAs in Mycoplasma pneumoniae cells | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Xue, L, Lenz, S, Rappsilber, J, Mahamid, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-05-25 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Visualizing translation dynamics at atomic detail inside a bacterial cell.
Nature, 610, 2022
|
|
7PIT
 
 | | 70S ribosome with EF-G, A/P- and P/E-site tRNAs in pseudouridimycin-treated Mycoplasma pneumoniae cells | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Xue, L, Lenz, S, Rappsilber, J, Mahamid, J. | | Deposit date: | 2021-08-23 | | Release date: | 2022-05-25 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Visualizing translation dynamics at atomic detail inside a bacterial cell.
Nature, 610, 2022
|
|
7PI8
 
 | | 70S ribosome with P-site tRNA in spectinomycin-treated Mycoplasma pneumoniae cells | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Xue, L, Lenz, S, Rappsilber, J, Mahamid, J. | | Deposit date: | 2021-08-19 | | Release date: | 2022-05-25 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Visualizing translation dynamics at atomic detail inside a bacterial cell.
Nature, 610, 2022
|
|
7PIS
 
 | | 70S ribosome with EF-G, A*- and P/E-site tRNAs in pseudouridimycin-treated Mycoplasma pneumoniae cells | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Xue, L, Lenz, S, Rappsilber, J, Mahamid, J. | | Deposit date: | 2021-08-23 | | Release date: | 2022-05-25 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Visualizing translation dynamics at atomic detail inside a bacterial cell.
Nature, 610, 2022
|
|
