4BS0
 
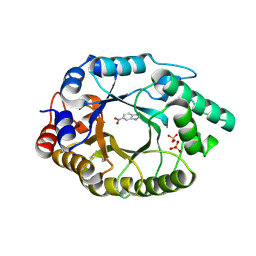 | | Crystal Structure of Kemp Eliminase HG3.17 E47N,N300D Complexed with Transition State Analog 6-Nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, KEMP ELIMINASE HG3.17, SULFATE ION | | Authors: | Blomberg, R, Kries, H, Pinkas, D.M, Mittl, P.R.E, Gruetter, M.G, Privett, H.K, Mayo, S, Hilvert, D. | | Deposit date: | 2013-06-06 | | Release date: | 2013-10-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Precision is Essential for Efficient Catalysis in an Evolved Kemp Eliminase
Nature, 503, 2013
|
|
2OKW
 
 | |
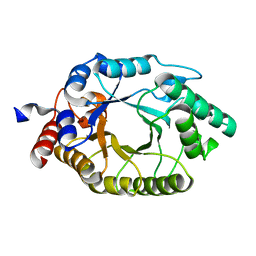
3O2L
 
 | |
3UAK
 
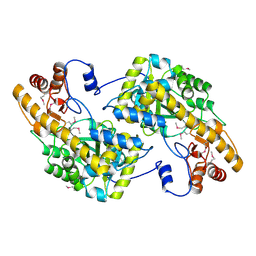 | | Crystal Structure of De Novo designed cysteine esterase ECH14, Northeast Structural Genomics Consortium Target OR54 | | Descriptor: | De Novo designed cysteine esterase ECH14 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Richter, F, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-10-21 | | Release date: | 2011-12-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.232 Å) | | Cite: | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
3U13
 
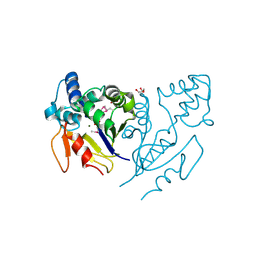 | | Crystal Structure of de Novo design of cystein esterase ECH13 at the resolution 1.6A, Northeast Structural Genomics Consortium Target OR51 | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Kohan, E, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-29 | | Release date: | 2011-11-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
3U1O
 
 | | THREE DIMENSIONAL STRUCTURE OF DE NOVO DESIGNED CYSTEINE ESTERASE ECH19, Northeast Structural Genomics Consortium Target OR49 | | Descriptor: | De Novo design cysteine esterase ECH19, SODIUM ION, SULFATE ION | | Authors: | Kuzin, A, Su, M, Lew, S, Forouhar, F, Seetharaman, J, Daya, P, Xiao, R, Ciccosanti, C, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-30 | | Release date: | 2011-10-26 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
3U1V
 
 | | X-ray Structure of De Novo design cysteine esterase FR29, Northeast Structural Genomics Consortium Target OR52 | | Descriptor: | De Novo design cysteine esterase FR29 | | Authors: | Kuzin, A, Su, M, Vorobiev, S.M, Seetharaman, J, Patel, D, Xiao, R, Ciccosanti, C, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-30 | | Release date: | 2011-12-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
2OKY
 
 | |
3NYD
 
 | | Crystal Structure of Kemp Eliminase HG-2 Complexed with Transition State Analog 5-Nitro Benzotriazole | | Descriptor: | 5-nitro-1H-benzotriazole, ACETATE ION, Endo-1,4-beta-xylanase, ... | | Authors: | Lee, T.M, Privett, H.K, Kaiser, J.T, Mayo, S.L. | | Deposit date: | 2010-07-14 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Iterative approach to computational enzyme design.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3NZ1
 
 | | Crystal Structure of Kemp Elimination Catalyst 1A53-2 Complexed with Transition State Analog 5-Nitro Benzotriazole | | Descriptor: | 5-nitro-1H-benzotriazole, Indole-3-glycerol phosphate synthase, L(+)-TARTARIC ACID, ... | | Authors: | Lee, T.M, Privett, H.K, Kaiser, J.T, Mayo, S.L. | | Deposit date: | 2010-07-15 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Iterative approach to computational enzyme design.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3NYZ
 
 | | Crystal Structure of Kemp Elimination Catalyst 1A53-2 | | Descriptor: | Indole-3-glycerol phosphate synthase, SULFATE ION | | Authors: | Lee, T.M, Privett, H.K, Kaiser, J.T, Mayo, S.L. | | Deposit date: | 2010-07-15 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.514 Å) | | Cite: | Iterative approach to computational enzyme design.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
