1ZCP
 
 | |
6X84
 
 | |
1A2J
 
 | | OXIDIZED DSBA CRYSTAL FORM II | | Descriptor: | DISULFIDE BOND FORMATION PROTEIN | | Authors: | Martin, J.L, Guddat, L.W. | | Deposit date: | 1998-01-06 | | Release date: | 1998-09-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of reduced and oxidized DsbA: investigation of domain motion and thiolate stabilization.
Structure, 6, 1998
|
|
1A2M
 
 | |
1A2L
 
 | | REDUCED DSBA AT 2.7 ANGSTROMS RESOLUTION | | Descriptor: | DISULFIDE BOND FORMATION PROTEIN | | Authors: | Martin, J.L, Guddat, L.W. | | Deposit date: | 1998-01-06 | | Release date: | 1998-07-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of reduced and oxidized DsbA: investigation of domain motion and thiolate stabilization.
Structure, 6, 1998
|
|
3DYR
 
 | |
1ACV
 
 | | DSBA MUTANT H32S | | Descriptor: | DSBA | | Authors: | Guddat, L.W, Martin, J.L. | | Deposit date: | 1997-02-10 | | Release date: | 1997-10-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of three His32 mutants of DsbA: support for an electrostatic role of His32 in DsbA stability.
Protein Sci., 6, 1997
|
|
1AC1
 
 | | DSBA MUTANT H32L | | Descriptor: | DSBA | | Authors: | Guddat, L.W, Martin, J.L. | | Deposit date: | 1997-02-10 | | Release date: | 1997-10-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of three His32 mutants of DsbA: support for an electrostatic role of His32 in DsbA stability.
Protein Sci., 6, 1997
|
|
5KCI
 
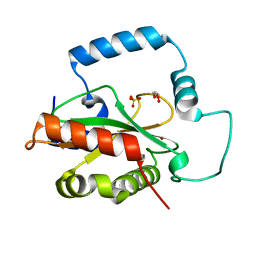 | | Crystal Structure of HTC1 | | Descriptor: | GLYCEROL, SULFATE ION, Uncharacterized protein YPL067C, ... | | Authors: | Martin, R.M, Horowitz, S, Koepnick, B, Cooper, S, Flatten, J, Rogawski, D.S, Koropatkin, N.M, Beinlich, F.R.M, Players, F, Students, U.M, Popovic, Z, Baker, D, Khatib, F, Bardwell, J.C.A. | | Deposit date: | 2016-06-06 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.833 Å) | | Cite: | Determining crystal structures through crowdsourcing and coursework.
Nat Commun, 7, 2016
|
|
1FVJ
 
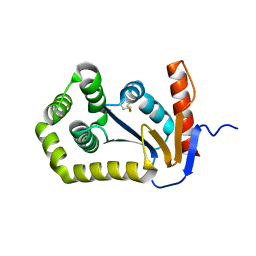 | |
1FVK
 
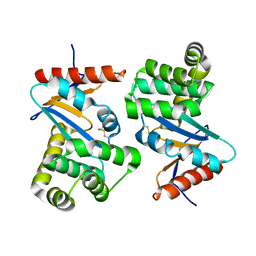 | |
1DSB
 
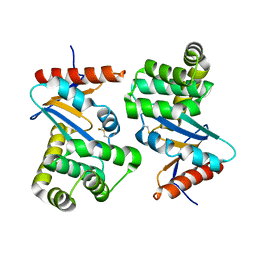 | |
3O39
 
 | |
6E0G
 
 | | Mitochondrial peroxiredoxin from Leishmania infantum after heat stress without unfolding client protein | | Descriptor: | mitochondrial 2-cys-peroxiredoxin | | Authors: | Teixeira, F, Tse, E, Makepeace, K.A.T, Borchers, C.H, Castro, H, Tomas, A.M, Poole, L.B, Southworth, D.R, Jakob, U. | | Deposit date: | 2018-07-06 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Chaperone activation and client binding of a 2-cysteine peroxiredoxin.
Nat Commun, 10, 2019
|
|
6E0F
 
 | | Mitochondrial peroxiredoxin from Leishmania infantum in complex with unfolding client protein after heat stress | | Descriptor: | mitochondrial 2-cys-peroxiredoxin | | Authors: | Teixeira, F, Tse, E, Makepeace, K.A.T, Borchers, C.H, Castro, H, Tomas, A.M, Poole, L.B, Southworth, D.R, Jakob, U. | | Deposit date: | 2018-07-06 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Chaperone activation and client binding of a 2-cysteine peroxiredoxin.
Nat Commun, 10, 2019
|
|
1DM9
 
 | | HEAT SHOCK PROTEIN 15 KD | | Descriptor: | HYPOTHETICAL 15.5 KD PROTEIN IN MRCA-PCKA INTERGENIC REGION, SULFATE ION | | Authors: | Staker, B.L, Korber, P, Bardwell, J.C.A, Saper, M.A. | | Deposit date: | 1999-12-14 | | Release date: | 2000-02-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Hsp15 reveals a novel RNA-binding motif.
EMBO J., 19, 2000
|
|
1EIZ
 
 | | FTSJ RNA METHYLTRANSFERASE COMPLEXED WITH S-ADENOSYLMETHIONINE | | Descriptor: | FTSJ, S-ADENOSYLMETHIONINE | | Authors: | Bugl, H, Fauman, E.B, Staker, B.L, Zheng, F, Kushner, S.R, Saper, M.A, Bardwell, J.C.A, Jakob, U. | | Deposit date: | 2000-02-29 | | Release date: | 2000-08-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | RNA methylation under heat shock control.
Mol.Cell, 6, 2000
|
|
1EJ0
 
 | | FTSJ RNA METHYLTRANSFERASE COMPLEXED WITH S-ADENOSYLMETHIONINE, MERCURY DERIVATIVE | | Descriptor: | FTSJ, MERCURY (II) ION, S-ADENOSYLMETHIONINE | | Authors: | Bugl, H, Fauman, E.B, Staker, B.L, Zheng, F, Kushner, S.R, Saper, M.A, Bardwell, J.C.A, Jakob, U. | | Deposit date: | 2000-02-29 | | Release date: | 2000-08-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | RNA methylation under heat shock control.
Mol.Cell, 6, 2000
|
|
5J7D
 
 | | Computationally Designed Thioredoxin dF106 | | Descriptor: | COPPER (II) ION, Designed Thioredoxin dF106 | | Authors: | Horowitz, S, Johansen, N, Olsen, J.G, Winther, J.R. | | Deposit date: | 2016-04-06 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Computational Redesign of Thioredoxin Is Hypersensitive toward Minor Conformational Changes in the Backbone Template.
J.Mol.Biol., 428, 2016
|
|
5WO1
 
 | | Chaperone Spy H96L bound to Im7 L18A L19A L37A (Im7 un-modeled) | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, IMIDAZOLE, ... | | Authors: | Horowitz, S, Koldewey, P, Martin, R, Bardwell, J.C.A. | | Deposit date: | 2017-08-01 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Visualizing chaperone-assisted protein folding.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5WO3
 
 | | Chaperone Spy bound to Im7 (Im7 un-modeled) | | Descriptor: | CHLORIDE ION, IMIDAZOLE, Periplasmic chaperone Spy, ... | | Authors: | Horowitz, S, Koldewey, P, Martin, R, Bardwell, J.C.A. | | Deposit date: | 2017-08-01 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Visualizing chaperone-assisted protein folding.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5WO2
 
 | | Chaperone Spy bound to Casein Fragment (Casein un-modeled) | | Descriptor: | CHLORIDE ION, IMIDAZOLE, Periplasmic chaperone Spy, ... | | Authors: | Horowitz, S, Koldewey, P, Martin, R, Bardwell, J.C.A. | | Deposit date: | 2017-08-01 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | Visualizing chaperone-assisted protein folding.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5WNW
 
 | | Chaperone Spy bound to Im7 6-45 ensemble | | Descriptor: | CHLORIDE ION, Colicin-E7 immunity protein, IMIDAZOLE, ... | | Authors: | Horowitz, S, Salmon, L, Koldewey, P, Ahlstrom, L.S, Martin, R, Xu, Q, Afonine, P.V, Trievel, R.C, Brooks, C.L, Bardwell, J.C.A. | | Deposit date: | 2017-08-01 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Visualizing chaperone-assisted protein folding.
Nat. Struct. Mol. Biol., 23, 2016
|
|
1BQ7
 
 | |
2JU5
 
 | | DsbH Oxidoreductase | | Descriptor: | Thioredoxin Disulfide Isomerase | | Authors: | Ulmer, T.S. | | Deposit date: | 2007-08-15 | | Release date: | 2007-11-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Insight into Disulfide Bond Catalysis in Chlamydia from the Structure and Function of DsbH, a Novel Oxidoreductase.
J.Biol.Chem., 283, 2008
|
|
