5M1W
 
 | | Structure of a stable G-hairpin | | Descriptor: | DNA (5'-D(*GP*TP*GP*TP*GP*GP*GP*TP*GP*TP*G)-3') | | Authors: | Gajarsky, M, Zivkovic, M.L, Stadlbauer, P, Pagano, B, Fiala, R, Amato, J, Tomaska, L, Sponer, J, Plavec, J, Trantirek, L. | | Deposit date: | 2016-10-11 | | Release date: | 2017-03-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a Stable G-Hairpin.
J. Am. Chem. Soc., 139, 2017
|
|
7PNL
 
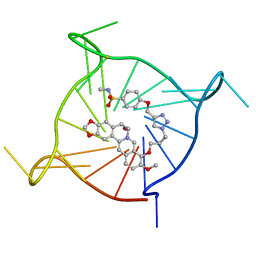 | | Complex between monomolecular human telomeric G-quadruplex and a sulfonamide derivative of the natural alkaloid Berberine | | Descriptor: | 4-[[1-[3-[(17-methoxy-5,7-dioxa-13-azoniapentacyclo[11.8.0.0^{2,10}.0^{4,8}.0^{15,20}]henicosa-1(21),2(10),3,8,13,15,17,19-octaen-16-yl)oxy]propyl]triazol-4-yl]methoxy]benzenesulfonamide, G-guadruplex DNA (23-mer), POTASSIUM ION | | Authors: | Bazzicalupi, C, Gratteri, P, Petreni, A, Nocentini, A. | | Deposit date: | 2021-09-07 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Development of a multi-targeted chemotherapeutic approach based on G-quadruplex stabilisation and carbonic anhydrase inhibition.
J Enzyme Inhib Med Chem, 39, 2024
|
|
2MKM
 
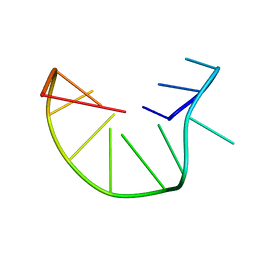 | | G-triplex structure and formation propensity | | Descriptor: | DNA_(5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*G)-3') | | Authors: | Cerofolini, L, Fragai, M, Giachetti, A, Limongelli, V, Luchinat, C, Novellino, E, Parrinello, M, Randazzo, A. | | Deposit date: | 2014-02-10 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | G-triplex structure and formation propensity.
Nucleic Acids Res., 42, 2014
|
|
2MKO
 
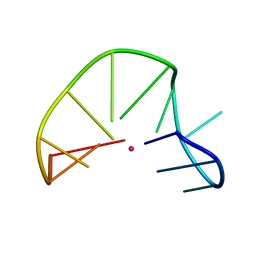 | | G-triplex structure and formation propensity | | Descriptor: | DNA_(5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*G)-3'), POTASSIUM ION | | Authors: | Cerofolini, L, Fragai, M, Giachetti, A, Limongelli, V, Luchinat, C, Novellino, E, Parrinello, M, Randazzo, A. | | Deposit date: | 2014-02-11 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | G-triplex structure and formation propensity.
Nucleic Acids Res., 42, 2014
|
|
6WI5
 
 | |
