3WA8
 
 | | Crystal structure of M. ruber CasB | | Descriptor: | CRISPR-associated protein, Cse2 family, MERCURY (II) ION | | Authors: | Yuan, Y.A, Yuan, Z. | | Deposit date: | 2013-04-28 | | Release date: | 2014-04-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into crRNA G-rich sequence binding and R-loop formation facilitated by Meiothermus ruber CasB
To be Published
|
|
5B1Z
 
 | |
5B4M
 
 | |
3W9U
 
 | | Crystal structure of Lipk107 | | Descriptor: | Putative lipase | | Authors: | Yuan, Y.A. | | Deposit date: | 2013-04-17 | | Release date: | 2013-12-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Lipk107
To be Published
|
|
3WTB
 
 | | Crystal structure of Gox0525 | | Descriptor: | Putative oxidoreductase | | Authors: | Yuan, Y.A, Lin, J.P. | | Deposit date: | 2014-04-09 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Gox0525
To be Published
|
|
3WBX
 
 | | Crystal structure of Gox0644 at apoform | | Descriptor: | Putative 2,5-diketo-D-gluconic acid reductase, SULFATE ION | | Authors: | Yuan, Y.A, Wang, C. | | Deposit date: | 2013-05-22 | | Release date: | 2014-05-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Gox0644 at apoform
TO BE PUBLISHED
|
|
3WBY
 
 | |
3WTC
 
 | | Crystal structure of Gox2036 | | Descriptor: | Putative oxidoreductase | | Authors: | Yuan, Y.A, Lin, J.P. | | Deposit date: | 2014-04-09 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of Gox0525
To be Published
|
|
3WJS
 
 | |
3AXJ
 
 | | High resolution crystal structure of C3PO | | Descriptor: | GM27569p, Translin associated factor X, isoform B | | Authors: | Yuan, Y.A, Yang, X. | | Deposit date: | 2011-04-07 | | Release date: | 2011-04-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High resolution crystal structure of C3PO
To be Published
|
|
3W1Y
 
 | |
3WBW
 
 | | Crystal structure of Gox0644 in complex with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative 2,5-diketo-D-gluconic acid reductase, SULFATE ION | | Authors: | Yuan, Y.A, Wang, C. | | Deposit date: | 2013-05-22 | | Release date: | 2014-05-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Gox0644 in complex with NADPH
To be Published
|
|
2RIM
 
 | | Crystal structure of Rtt109 | | Descriptor: | Regulator of Ty1 transposition protein 109 | | Authors: | Yuan, Y.A. | | Deposit date: | 2007-10-12 | | Release date: | 2008-09-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into histone h3 lysine 56 acetylation by rtt109
Structure, 16, 2008
|
|
3VZ3
 
 | | Structural insights into substrate and cofactor selection by sp2771 | | Descriptor: | 4-oxobutanoic acid, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Succinate-semialdehyde dehydrogenase | | Authors: | Yuan, Y.A, Yuan, Z, Yin, B, Wei, D. | | Deposit date: | 2012-10-09 | | Release date: | 2013-07-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural basis for cofactor and substrate selection by cyanobacterium succinic semialdehyde dehydrogenase
J.Struct.Biol., 182, 2013
|
|
3VZ1
 
 | |
3VZ0
 
 | | Structural insights into cofactor and substrate selection by Gox0499 | | Descriptor: | NONAETHYLENE GLYCOL, Putative NAD-dependent aldehyde dehydrogenase | | Authors: | Yuan, Y.A, Yuan, Z, Yin, B, Wei, D. | | Deposit date: | 2012-10-09 | | Release date: | 2013-07-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for cofactor and substrate selection by cyanobacterium succinic semialdehyde dehydrogenase
J.Struct.Biol., 182, 2013
|
|
3WJ7
 
 | | Crystal structure of gox2253 | | Descriptor: | MERCURY (II) ION, Putative oxidoreductase | | Authors: | Yuan, Y.A, Yin, B. | | Deposit date: | 2013-10-05 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into substrate and coenzyme preference by SDR family protein Gox2253 from Gluconobater oxydans.
Proteins, 82, 2014
|
|
3WJM
 
 | | Crystal structure of Bombyx mori Sp2/Sp3 heterohexamer | | Descriptor: | Arylphorin, Silkworm storage protein, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yuan, Y.A, Hou, Y. | | Deposit date: | 2013-10-11 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Bombyx mori arylphorins reveals a 3:3 heterohexamer with multiple papain cleavage sites
Protein Sci., 23, 2014
|
|
3WUY
 
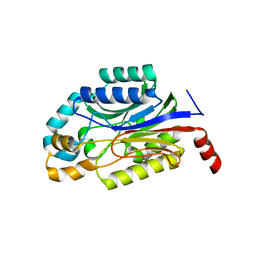 | | Crystal structure of Nit6803 | | Descriptor: | Nitrilase | | Authors: | Yuan, Y.A, Yin, B, Wang, C. | | Deposit date: | 2014-05-09 | | Release date: | 2014-12-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural insights into enzymatic activity and substrate specificity determination by a single amino acid in nitrilase from Syechocystis sp. PCC6803
J.Struct.Biol., 188, 2014
|
|
3WVO
 
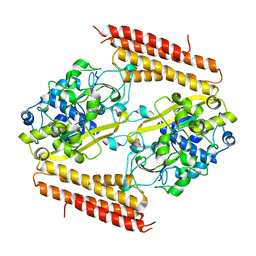 | | Crystal structure of Thermobifida fusca Cse1 | | Descriptor: | CRISPR-associated protein, Cse1 family | | Authors: | Yuan, Y.A, Tay, M. | | Deposit date: | 2014-06-02 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystal structure of Thermobifida fusca Cse1 reveals target DNA binding site.
Protein Sci., 24, 2015
|
|
3WZG
 
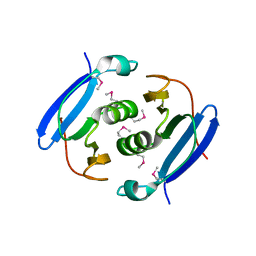 | | Crystal structure of AfCsx3 | | Descriptor: | Uncharacterized protein AF_1864 | | Authors: | Yuan, Y.A, Yan, X. | | Deposit date: | 2014-09-25 | | Release date: | 2015-07-15 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structures of CRISPR-associated Csx3 reveal a manganese-dependent deadenylation exoribonuclease.
Rna Biol., 12, 2015
|
|
3WZI
 
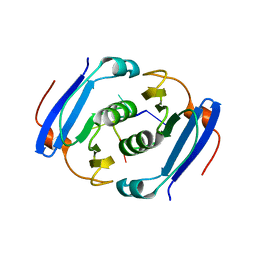 | | Crystal structure of AfCsx3 in complex with ssRNA | | Descriptor: | Uncharacterized protein AF_1864, ssRNA | | Authors: | Yuan, Y.A, Yan, X. | | Deposit date: | 2014-09-25 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of CRISPR-associated Csx3 reveal a manganese-dependent deadenylation exoribonuclease.
Rna Biol., 12, 2015
|
|
3WZH
 
 | | Crystal structure of AfCsx3 | | Descriptor: | MANGANESE (II) ION, Uncharacterized protein AF_1864 | | Authors: | Yuan, Y.A, Yan, X. | | Deposit date: | 2014-09-25 | | Release date: | 2015-07-15 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystal structures of CRISPR-associated Csx3 reveal a manganese-dependent deadenylation exoribonuclease.
Rna Biol., 12, 2015
|
|
2ZI0
 
 | | Crystal structure of Tav2b/siRNA complex | | Descriptor: | Protein 2b, RNA (5'-D(P*AP*GP*AP*CP*AP*GP*CP*AP*UP*UP*AP*UP*GP*CP*UP*GP*UP*CP*UP*UP*U)-3') | | Authors: | Yuan, Y.A, Chen, H.-Y. | | Deposit date: | 2008-02-12 | | Release date: | 2008-07-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural basis for RNA-silencing suppression by Tomato aspermy virus protein 2b
Embo Rep., 9, 2008
|
|
2ZFN
 
 | |
