1F2D
 
 | | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE DEAMINASE | | Descriptor: | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE DEAMINASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Yao, M, Ose, T, Sugimoto, H, Horiuchi, A, Nakagawa, A, Yokoi, D, Murakami, T, Honma, M, Wakatsuki, S, Tanaka, I. | | Deposit date: | 2000-05-24 | | Release date: | 2000-12-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 1-aminocyclopropane-1-carboxylate deaminase from Hansenula saturnus.
J.Biol.Chem., 275, 2000
|
|
1IZ6
 
 | | Crystal Structure of Translation Initiation Factor 5A from Pyrococcus Horikoshii | | Descriptor: | Initiation Factor 5A | | Authors: | Yao, M, Ohsawa, A, Kikukawa, S, Tanaka, I, Kimura, M. | | Deposit date: | 2002-09-25 | | Release date: | 2003-01-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Hyperthermophilic Archaeal Initiation Factor 5A: A Homologue of Eukaryotic Initiation Factor 5A (eIF-5A)
J.BIOCHEM.(TOKYO), 133, 2003
|
|
1WLS
 
 | | Crystal structure of L-asparaginase I homologue protein from Pyrococcus horikoshii | | Descriptor: | L-asparaginase | | Authors: | Yao, M, Morita, H, Yasutake, Y, Tanaka, I. | | Deposit date: | 2004-06-29 | | Release date: | 2005-03-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure of the type I L-asparaginase from the hyperthermophilic archaeon Pyrococcus horikoshii at 2.16 angstroms resolution.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1V7L
 
 | |
1VGJ
 
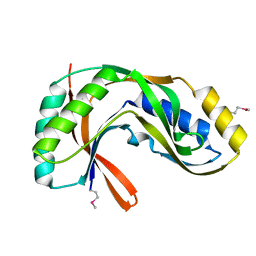 | | Crystal structure of 2'-5' RNA ligase from Pyrococcus horikoshii | | Descriptor: | Hypothetical protein PH0099 | | Authors: | Yao, M, Morita, H, Okada, A, Tanaka, I. | | Deposit date: | 2004-04-27 | | Release date: | 2005-06-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The structure of Pyrococcus horikoshii 2'-5' RNA ligase at 1.94 A resolution reveals a possible open form with a wider active-site cleft
ACTA CRYSTALLOGR.,SECT.F, 62, 2006
|
|
2ZQ0
 
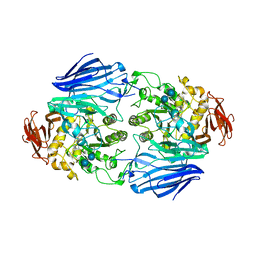 | | Crystal structure of SusB complexed with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glucosidase (Alpha-glucosidase SusB), CALCIUM ION | | Authors: | Yao, M, Tanaka, I, Kitamura, M. | | Deposit date: | 2008-07-31 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional analysis of a glycoside hydrolase family 97 enzyme from Bacteroides thetaiotaomicron.
J.Biol.Chem., 283, 2008
|
|
1QQY
 
 | |
5XFM
 
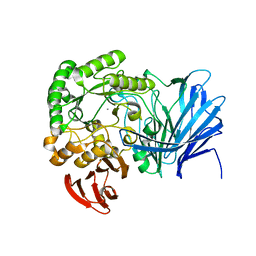 | | Crystal structure of beta-arabinopyranosidase | | Descriptor: | Alpha-glucosidase, CALCIUM ION | | Authors: | Kato, K, Okuyama, M, Yao, M. | | Deposit date: | 2017-04-10 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel glycoside hydrolase family 97 enzyme: Bifunctional beta-l-arabinopyranosidase/ alpha-galactosidase from Bacteroides thetaiotaomicron.
Biochimie, 142, 2017
|
|
5X3I
 
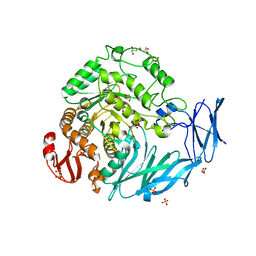 | | Kfla1895 D451A mutant | | Descriptor: | GLYCEROL, Glycoside hydrolase family 31, SULFATE ION | | Authors: | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | Deposit date: | 2017-02-06 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Glycoside hydrolase mutant
To Be Published
|
|
5X3J
 
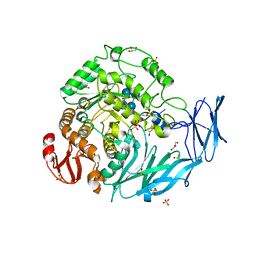 | | Kfla1895 D451A mutant in complex with cyclobis-(1->6)-alpha-nigerosyl | | Descriptor: | Cyclic alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose, GLYCEROL, Glycoside hydrolase family 31, ... | | Authors: | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | Deposit date: | 2017-02-06 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Glycoside hydrolase mutant in complex with substrate
To Be Published
|
|
5X6C
 
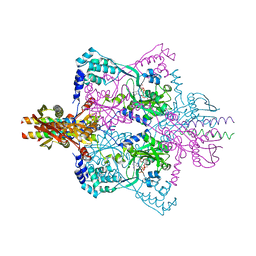 | | Crystal structure of SepRS-SepCysE from Methanocaldococcus jannaschii | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, O-phosphoserine--tRNA(Cys) ligase, SULFATE ION, ... | | Authors: | Chen, M, Kato, K, Yao, M. | | Deposit date: | 2017-02-21 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Structural basis for tRNA-dependent cysteine biosynthesis
Nat Commun, 8, 2017
|
|
5X6B
 
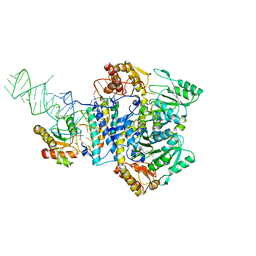 | |
5X32
 
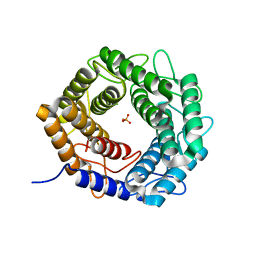 | | Crystal structure of D-mannose isomerase | | Descriptor: | N-acylglucosamine 2-epimerase, PHOSPHATE ION | | Authors: | Kato, K, Saburi, W, Yao, M. | | Deposit date: | 2017-02-03 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Biochemical and structural characterization of Marinomonas mediterranead-mannose isomerase Marme_2490 phylogenetically distant from known enzymes
Biochimie, 144, 2018
|
|
4WLC
 
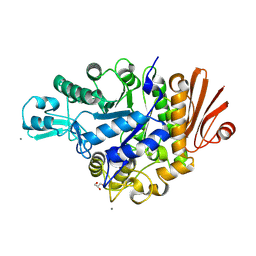 | | Structure of dextran glucosidase with glucose | | Descriptor: | CALCIUM ION, GLYCEROL, Glucan 1,6-alpha-glucosidase, ... | | Authors: | Kobayashi, M, Kato, K, Yao, M. | | Deposit date: | 2014-10-07 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Structural insights into the catalytic reaction that is involved in the reorientation of Trp238 at the substrate-binding site in GH13 dextran glucosidase
Febs Lett., 589, 2015
|
|
4XB3
 
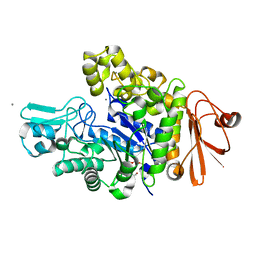 | | Structure of dextran glucosidase | | Descriptor: | CALCIUM ION, Glucan 1,6-alpha-glucosidase, HEXAETHYLENE GLYCOL | | Authors: | Kobayashi, M, Kato, K, Yao, M. | | Deposit date: | 2014-12-16 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Structural insights into the catalytic reaction that is involved in the reorientation of Trp238 at the substrate-binding site in GH13 dextran glucosidase
Febs Lett., 589, 2015
|
|
4V60
 
 | | The structure of rat liver vault at 3.5 angstrom resolution | | Descriptor: | Major vault protein | | Authors: | Kato, K, Zhou, Y, Tanaka, H, Yao, M, Yamashita, E, Yoshimura, M, Tsukihara, T. | | Deposit date: | 2008-10-24 | | Release date: | 2014-07-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of rat liver vault at 3.5 angstrom resolution
Science, 323, 2009
|
|
3RXD
 
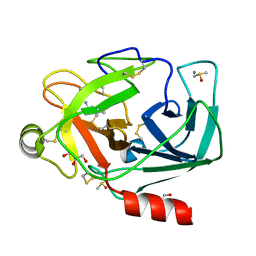 | | Crystal structure of Trypsin complexed with (3-methoxyphenyl)methanamine | | Descriptor: | 1-(3-methoxyphenyl)methanamine, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXB
 
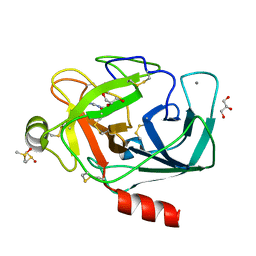 | | Crystal structure of Trypsin complexed with 4-guanidinobutanoic acid | | Descriptor: | 4-carbamimidamidobutanoic acid, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXP
 
 | | Crystal structure of Trypsin complexed with (1,5-dimethylpyrazol-3-yl)methanamine | | Descriptor: | 1-(1,5-dimethyl-1H-pyrazol-3-yl)methanamine, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXM
 
 | | Crystal structure of Trypsin complexed with [2-(2-thienyl)thiazol-4-yl]methanamine | | Descriptor: | 1-[2-(thiophen-2-yl)-1,3-thiazol-4-yl]methanamine, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXH
 
 | | Crystal structure of Trypsin complexed with 2-(1H-imidazol-4-yl)ethanamine | | Descriptor: | CALCIUM ION, Cationic trypsin, DIMETHYL SULFOXIDE, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXF
 
 | | Crystal structure of Trypsin complexed with 4-aminopyridine | | Descriptor: | 4-AMINOPYRIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXG
 
 | | Crystal structure of Trypsin complexed with 4-aminocyclohexanol | | Descriptor: | CALCIUM ION, Cationic trypsin, DIMETHYL SULFOXIDE, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXT
 
 | | Crystal structure of Trypsin complexed with (3-methoxyphenyl)methanamin (F04 and F03, cocktail experiment) | | Descriptor: | 1-(3-methoxyphenyl)methanamine, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXC
 
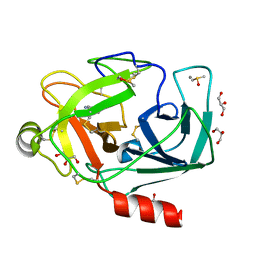 | | Crystal structure of Trypsin complexed with 2-aminopyridine | | Descriptor: | 2-AMINOPYRIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
