2IIH
 
 | | Crystal structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from thermus theromophilus HB8 (H32 form) | | Descriptor: | Molybdenum cofactor biosynthesis protein C, PHOSPHATE ION | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, Baba, S, Chen, L, Liu, Z.-J, Wang, B.-C, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-28 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from thermus theromophilus HB8 (H32 form)
To be Published
|
|
1SL9
 
 | | Obelin from Obelia longissima | | Descriptor: | C2-HYDROPEROXY-COELENTERAZINE, Obelin | | Authors: | Deng, L, Markova, S, Vysotski, E, Liu, Z.-J, Lee, J, Rose, J, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-03-05 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Obelin from Obelia longissima
To be Published
|
|
1S36
 
 | | Crystal structure of a Ca2+-discharged photoprotein: Implications for the mechanisms of the calcium trigger and the bioluminescence | | Descriptor: | CHLORIDE ION, GLYCEROL, N-[3-BENZYL-5-(4-HYDROXYPHENYL)PYRAZIN-2-YL]-2-(4-HYDROXYPHENYL)ACETAMIDE, ... | | Authors: | Deng, L, Markova, S.V, Vysotski, E.S, Liu, Z.-J, Lee, J, Rose, J, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-01-12 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of a Ca2+-discharged photoprotein: implications for mechanisms of the calcium trigger and bioluminescence
J.Biol.Chem., 279, 2004
|
|
2PH7
 
 | | Crystal structure of AF2093 from Archaeoglobus fulgidus | | Descriptor: | Uncharacterized protein AF_2093 | | Authors: | Chang, J.C, Yang, H, Hwang, J, Zhu, J, Chen, L, Fu, Z.-Q, Xu, H, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2007-04-10 | | Release date: | 2007-05-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of AF2093 from Archaeoglobus fulgidus.
To be Published
|
|
3E19
 
 | | Crystal Structure of Iron Uptake Regulatory Protein (FeoA) Solved by Sulfur SAD in a Monoclinic Space Group | | Descriptor: | FeoA, GLYCEROL, PHOSPHATE ION | | Authors: | Hughes, R.C, Li, Y, Wang, B.-C, Liu, Z.-J, Ng, J.D. | | Deposit date: | 2008-08-02 | | Release date: | 2008-12-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic Structure Determination of Iron Uptake Regulatory
Protein (FeoA) by Sulfur SAD in a Monoclinic Space Group
To be Published
|
|
2Z0M
 
 | | Crystal structure of hypothetical ATP-dependent RNA helicase from Sulfolobus tokodaii | | Descriptor: | 337aa long hypothetical ATP-dependent RNA helicase deaD | | Authors: | Nakagawa, N, Kusano, S, Shirouzu, M, Chen, L, Fu, Z.-Q, Chrzas, J, Wang, B.-C, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of hypothetical ATP-dependent RNA helicase from Sulfolobus tokodaii
To be Published
|
|
1JF0
 
 | | The Crystal Structure of Obelin from Obelia geniculata at 1.82 A Resolution | | Descriptor: | C2-HYDROPEROXY-COELENTERAZINE, Obelin | | Authors: | Deng, L, Vysotski, E, Liu, Z.-J, Markova, S, Lee, J, Rose, J, Wang, B.-C. | | Deposit date: | 2001-06-19 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Crystal Structure of Obelin from Obelia geniculata at 1.82 Angstrom Resolution
To be Published
|
|
1OQC
 
 | |
1ZTD
 
 | | Hypothetical Protein Pfu-631545-001 From Pyrococcus furiosus | | Descriptor: | Hypothetical Protein Pfu-631545-001 | | Authors: | Fu, Z.-Q, Horanyi, P, Florence, Q, Liu, Z.-J, Chen, L, Lee, D, Habel, J, Xu, H, Nguyen, D, Chang, S.-H, Zhou, W, Zhang, H, Jenney Jr, F.E, Sha, B, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-05-26 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hypothetical Protein Pfu-631545-001 From Pyrococcus furiosus
To be Published
|
|
2A4K
 
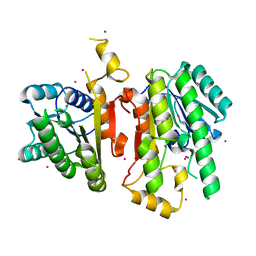 | | 3-Oxoacyl-[acyl carrier protein] reductase from Thermus thermophilus TT0137 | | Descriptor: | 3-oxoacyl-[acyl carrier protein] reductase, UNKNOWN ATOM OR ION | | Authors: | Zhou, W, Ebihara, A, Tempel, W, Yokoyama, S, Chen, L, Kuramitsu, S, Nguyen, J, Chang, S.-H, Liu, Z.-J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-29 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | 3-Oxoacyl-[acyl carrier protein] reductase from Thermus thermophilus TT0137
To be Published
|
|
1JF2
 
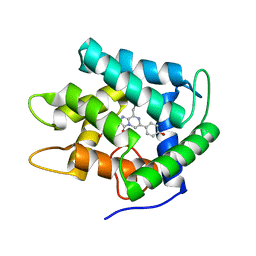 | | Crystal Structure of W92F obelin mutant from Obelia longissima at 1.72 Angstrom resolution | | Descriptor: | C2-HYDROPEROXY-COELENTERAZINE, obelin | | Authors: | Liu, Z.-J, Vysotski, E.S, Deng, L, Markova, S.V, Lee, J, Rose, J.P, Wang, B.-C. | | Deposit date: | 2001-06-19 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Violet bioluminescence and fast kinetics from W92F obelin: structure-based proposals for the bioluminescence triggering and the identification of the emitting species.
Biochemistry, 42, 2003
|
|
1GSC
 
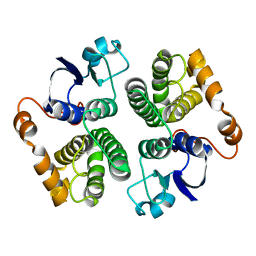 | |
1GSB
 
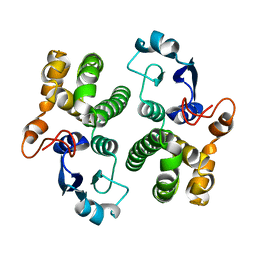 | |
3KPX
 
 | | Crystal Structure Analysis of photoprotein clytin | | Descriptor: | Apophotoprotein clytin-3, C2-HYDROPEROXY-COELENTERAZINE, CALCIUM ION | | Authors: | Titushin, M.S, Li, Y, Stepanyuk, G.A, Wang, B.-C, Lee, J, Vysotski, E.S, Liu, Z.-J. | | Deposit date: | 2009-11-17 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | NMR derived topology of a GFP-photoprotein energy transfer complex
J.Biol.Chem., 285, 2010
|
|
1NB2
 
 | | Crystal Structure of Nucleoside Diphosphate Kinase from Bacillus Halodenitrificans | | Descriptor: | Nucleoside Diphosphate Kinase | | Authors: | Chen, C.-J, Liu, M.-Y, Chang, W.-C, Chang, T, Wang, B.-C, Le Gall, J. | | Deposit date: | 2002-12-02 | | Release date: | 2003-05-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a nucleoside diphosphate kinase from Bacillus halodenitrificans: coexpression of its activity with a Mn-superoxide dismutase.
J.Struct.Biol., 142, 2003
|
|
1LPL
 
 | | Structural Genomics of Caenorhabditis elegans: CAP-Gly domain of F53F4.3 | | Descriptor: | Hypothetical 25.4 kDa protein F53F4.3 in chromosome V | | Authors: | Li, S, Finley, J, Liu, Z.-J, Qiu, S.H, Luan, C.H, Carson, M, Tsao, J, Johnson, D, Lin, G, Zhao, J, Thomas, W, Nagy, L.A, Sha, B, DeLucas, L.J, Wang, B.-C, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2002-05-08 | | Release date: | 2002-05-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal Structure of the
Cytoskeleton-associated Protein
Glycine-rich (CAP-Gly) Domain
J.Biol.Chem., 277, 2002
|
|
5UVM
 
 | | HIT family hydrolase from Clostridium thermocellum Cth-393 | | Descriptor: | ADENOSINE, Histidine triad (HIT) protein, UNKNOWN ATOM OR ION, ... | | Authors: | Habel, J, Tempel, W, Liu, Z.-J, Rose, J, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2017-02-20 | | Release date: | 2017-04-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | HIT family hydrolase from Clostridium thermocellum Cth-393
To Be Published
|
|
1NNQ
 
 | | rubrerythrin from Pyrococcus furiosus Pfu-1210814 | | Descriptor: | Rubrerythrin, ZINC ION | | Authors: | Liu, Z.-J, Tempel, W, Schubot, F.D, Shah, A, Arendall III, W.B, Rose, J.P, Richardson, D.C, Richardson, J.S, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-01-14 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural genomics of Pyrococcus furiosus: X-ray crystallography reveals 3D domain swapping in rubrerythrin
Proteins, 57, 2004
|
|
1PRY
 
 | | Structure Determination of Fibrillarin Homologue From Hyperthermophilic Archaeon Pyrococcus furiosus (Pfu-65527) | | Descriptor: | Fibrillarin-like pre-rRNA processing protein | | Authors: | Deng, L, Starostina, N.G, Liu, Z.-J, Rose, J.P, Terns, R.M, Terns, M.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-06-20 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure determination of fibrillarin from the hyperthermophilic archaeon Pyrococcus furiosus
Biochem.Biophys.Res.Commun., 315, 2004
|
|
1NXC
 
 | | Structure of mouse Golgi alpha-1,2-mannosidase IA reveals the molecular basis for substrate specificity among Class I enzymes (family 47 glycosidases) | | Descriptor: | CALCIUM ION, Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA, alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Tempel, W, Liu, Z.-J, Karaveg, K, Rose, J, Moremen, K.W, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-02-10 | | Release date: | 2004-05-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure of mouse Golgi alpha-mannosidase IA reveals the molecular basis for substrate specificity among class 1 (family 47 glycosylhydrolase) alpha1,2-mannosidases
J.Biol.Chem., 279, 2004
|
|
2DUY
 
 | | Crystal structure of competence protein ComEA-related protein from Thermus thermophilus HB8 | | Descriptor: | CHLORIDE ION, Competence protein ComEA-related protein | | Authors: | Niwa, H, Shimada, A, Chen, L, Liu, Z.-J, Wang, B.-C, Ebihara, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-07-27 | | Release date: | 2007-08-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of competence protein ComEA-related protein from Thermus thermophilus HB8
To be Published
|
|
2FZF
 
 | | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus | | Descriptor: | hypothetical protein | | Authors: | Fu, Z.-Q, Liu, Z.-J, Lee, D, Kelley, L, Chen, L, Tempel, W, Shah, N, Horanyi, P, Lee, H.S, Habel, J, Dillard, B.D, Nguyen, D, Chang, S.-H, Zhang, H, Chang, J, Sugar, F.J, Poole, F.L, Jenney Jr, F.E, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-02-09 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus
To be published
|
|
2EFK
 
 | | Crystal structure of the EFC domain of Cdc42-interacting protein 4 | | Descriptor: | Cdc42-interacting protein 4 | | Authors: | Shimada, A, Niwa, H, Chen, L, Liu, Z.-J, Wang, B.-C, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-23 | | Release date: | 2007-05-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Curved EFC/F-BAR-Domain Dimers Are Joined End to End into a Filament for Membrane Invagination in Endocytosis
Cell(Cambridge,Mass.), 129, 2007
|
|
2EIS
 
 | | X-ray structure of acyl-CoA hydrolase-like protein, TT1379, from Thermus thermophilus HB8 | | Descriptor: | COENZYME A, Hypothetical protein TTHB207 | | Authors: | Kamitori, S, Yoshida, H, Satoh, S, Iino, H, Ebihara, A, Chen, L, Fu, Z.-Q, Chrzas, J, Wang, B.-C, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-13 | | Release date: | 2008-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structure of acyl-CoA hydrolase-like protein, TT1379, from Thermus thermophilus HB8
To be Published
|
|
2HIQ
 
 | | Crystal structure of JW1657 from Escherichia coli | | Descriptor: | Hypothetical protein ydhR | | Authors: | Chen, L.Q, Chen, L.R, Liu, Z.-J, Temple, W, Lee, D, Chang, S.-H, Rose, J.P, Ebihara, A, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-06-29 | | Release date: | 2006-09-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of JW1657 from Escherichia coli at 2.0A resolution
To be Published
|
|
