1NI0
 
 | |
1ADU
 
 | | EARLY E2A DNA-BINDING PROTEIN | | Descriptor: | ADENOVIRUS SINGLE-STRANDED DNA-BINDING PROTEIN, ZINC ION | | Authors: | Tucker, P.A, Kanellopoulos, P.N, Tsernoglou, D, Van Der Vliet, P.C. | | Deposit date: | 1995-05-11 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Alternative arrangements of the protein chain are possible for the adenovirus single-stranded DNA binding protein.
J.Mol.Biol., 257, 1996
|
|
1L0Z
 
 | | THE STRUCTURE OF PORCINE PANCREATIC ELASTASE COMPLEXED WITH XENON AND BROMIDE, CRYOPROTECTED WITH DRY PARAFFIN OIL | | Descriptor: | BROMIDE ION, ELASTASE 1, SODIUM ION, ... | | Authors: | Tucker, P.A, Panjikar, S. | | Deposit date: | 2002-02-14 | | Release date: | 2002-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Xenon derivatization of halide-soaked protein crystals.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3K3C
 
 | |
3ETI
 
 | |
3EW5
 
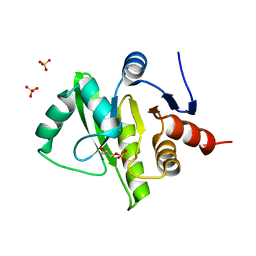 | | Structure of the tetragonal crystal form of X (ADRP) domain from FCoV | | Descriptor: | CHLORIDE ION, SN-GLYCEROL-1-PHOSPHATE, SULFATE ION, ... | | Authors: | Wojdyla, J.A, Manolaridis, I, Tucker, P.A. | | Deposit date: | 2008-10-14 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the X (ADRP) domain of nsp3 from feline coronavirus
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3KE6
 
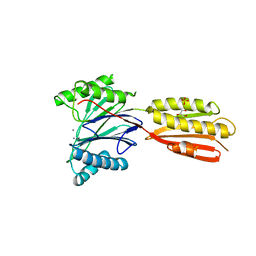 | |
3MP2
 
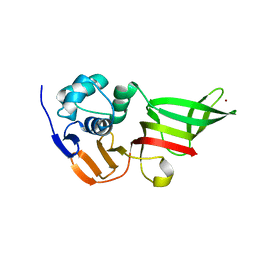 | |
3K3D
 
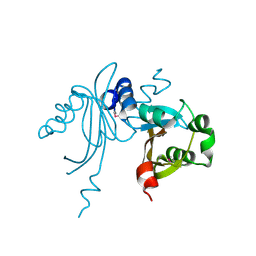 | |
3GZF
 
 | | Structure of the C-terminal domain of nsp4 from Feline Coronavirus | | Descriptor: | Replicase polyprotein 1ab, SULFATE ION | | Authors: | Manolaridis, I, Wojdyla, J.A, Panjikar, S, Snijder, E.J, Gorbalenya, A.E, Coutard, B, Tucker, P.A. | | Deposit date: | 2009-04-07 | | Release date: | 2009-08-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.756 Å) | | Cite: | Structure of the C-terminal domain of nsp4 from feline coronavirus
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3JZT
 
 | | Structure of a cubic crystal form of X (ADRP) domain from FCoV with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHLORIDE ION, SODIUM ION, ... | | Authors: | Wojdyla, J.A, Manolaridis, I, Tucker, P.A. | | Deposit date: | 2009-09-24 | | Release date: | 2010-01-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.91 Å) | | Cite: | Structure of the X (ADRP) domain of nsp3 from feline coronavirus
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2YKH
 
 | | Sensor region of a sensor histidine kinase | | Descriptor: | PROBABLE SENSOR HISTIDINE KINASE PDTAS | | Authors: | Preu, J, Panjikar, S, Morth, P, Jaiswal, R, Karunakar, P, Tucker, P.A. | | Deposit date: | 2011-05-27 | | Release date: | 2012-06-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | The Sensor Region of the Ubiquitous Cytosolic Sensor Kinase, Pdtas, Contains Pas and Gaf Domain Sensing Modules.
J.Struct.Biol., 177, 2012
|
|
1ANV
 
 | | ADENOVIRUS 5 DBP/URANYL FLUORIDE SOAK | | Descriptor: | ADENOVIRUS SINGLE-STRANDED DNA-BINDING PROTEIN, URANYL (VI) ION, ZINC ION | | Authors: | Kanellopoulos, P.N, Tsernoglou, D, Van Der Vliet, P.C, Tucker, P.A. | | Deposit date: | 1996-03-14 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational change of the adenovirus DNA-binding protein induced by soaking crystals with K3UO2F5 solutions.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1VLN
 
 | |
1UVO
 
 | | Structure Of The Complex Of Porcine Pancreatic Elastase In Complex With Cadmium Refined At 1.85 A Resolution (Crystal A) | | Descriptor: | ACETATE ION, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Weiss, M.S, Panjikar, S, Mueller-Dieckmann, C, Tucker, P.A. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | On the Influence of the Incident Photon Energy on the Radiation Damage in Crystalline Biological Samples
J.Synchrotron Radiat., 12, 2005
|
|
1UO6
 
 | | PORCINE PANCREATIC ELASTASE/Xe-COMPLEX | | Descriptor: | CHLORIDE ION, ELASTASE 1, GLYCEROL, ... | | Authors: | Mueller-Dieckmann, C, Polentarutti, M, Djinovic-Carugo, K, Panjikar, S, Tucker, P.A, Weiss, M.S. | | Deposit date: | 2003-09-16 | | Release date: | 2003-10-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | On the Routine Use of Soft X-Rays in Macromolecular Crystallography. Part II. Data-Collection Wavelength and Scaling Models
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1UVP
 
 | | Structure Of The Complex Of Porcine Pancreatic Elastase In Complex With Cadmium Refined At 1.85 A Resolution (Crystal B) | | Descriptor: | ACETATE ION, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Weiss, M.S, Panjikar, S, Mueller-Dieckmann, C, Tucker, P.A. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | On the Influence of the Incident Photon Energy on the Radiation Damage in Crystalline Biological Samples
J.Synchrotron Radiat., 12, 2005
|
|
1URJ
 
 | | Single stranded DNA-binding protein(ICP8) from Herpes simplex virus-1 | | Descriptor: | MAJOR DNA-BINDING PROTEIN, MERCURY (II) ION, ZINC ION | | Authors: | Panjikar, S, Mapelli, M, Tucker, P.A. | | Deposit date: | 2003-10-30 | | Release date: | 2004-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of the herpes simplex virus 1 ssDNA-binding protein suggests the structural basis for flexible, cooperative single-stranded DNA binding.
J. Biol. Chem., 280, 2005
|
|
1VAL
 
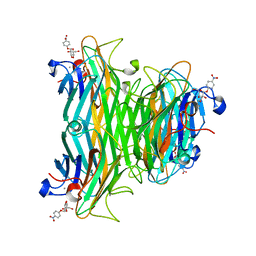 | | CONCANAVALIN A COMPLEX WITH 4'-NITROPHENYL-ALPHA-D-GLUCOPYRANOSIDE | | Descriptor: | 4-nitrophenyl alpha-D-glucopyranoside, CALCIUM ION, CONCANAVALIN A, ... | | Authors: | Kanellopoulos, P.N, Tucker, P.A, Hamodrakas, S.J. | | Deposit date: | 1996-01-08 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of the complexes of concanavalin A with 4'-nitrophenyl-alpha-D-mannopyranoside and 4'-nitrophenyl-alpha-D-glucopyranoside.
J.Struct.Biol., 116, 1996
|
|
1VAM
 
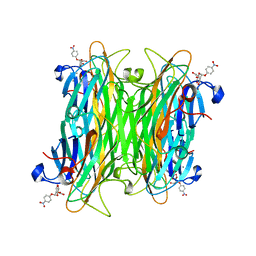 | | CONCANAVALIN A COMPLEX WITH 4'-NITROPHENYL-ALPHA-D-MANNOPYRANOSIDE | | Descriptor: | 4-nitrophenyl alpha-D-mannopyranoside, CALCIUM ION, CONCANAVALIN A, ... | | Authors: | Kanellopoulos, P.N, Tucker, P.A, Hamodrakas, S.J. | | Deposit date: | 1996-01-08 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The crystal structure of the complexes of concanavalin A with 4'-nitrophenyl-alpha-D-mannopyranoside and 4'-nitrophenyl-alpha-D-glucopyranoside.
J.Struct.Biol., 116, 1996
|
|
1ADV
 
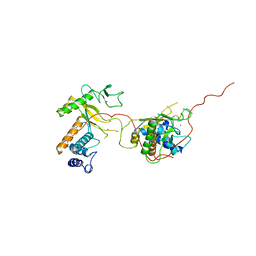 | | EARLY E2A DNA-BINDING PROTEIN | | Descriptor: | ADENOVIRUS SINGLE-STRANDED DNA-BINDING PROTEIN, ZINC ION | | Authors: | Kanellopoulos, P.N, Tsernoglou, D, Van Der Vliet, P.C, Tucker, P.A. | | Deposit date: | 1995-05-12 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Alternative arrangements of the protein chain are possible for the adenovirus single-stranded DNA binding protein.
J.Mol.Biol., 257, 1996
|
|
1CJP
 
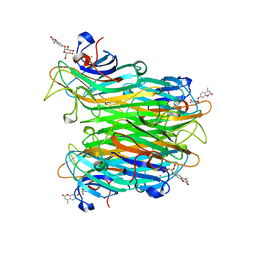 | | CONCANAVALIN A COMPLEX WITH 4'-METHYLUMBELLIFERYL-ALPHA-D-GLUCOPYRANOSIDE | | Descriptor: | 4-METHYLUMBELLIFERYL-ALPHA-D-GLUCOSE, CALCIUM ION, CONCANAVALIN A, ... | | Authors: | Hamodrakas, S.J, Kanellopoulos, P.N, Tucker, P.A. | | Deposit date: | 1996-10-03 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | The crystal structure of the complex of concanavalin A with 4'-methylumbelliferyl-alpha-D-glucopyranoside.
J.Struct.Biol., 118, 1997
|
|
1ARL
 
 | | CARBOXYPEPTIDASE A WITH ZN REMOVED | | Descriptor: | APO-CARBOXYPEPTIDASE A=ALPHA= (COX) | | Authors: | Greenblatt, H.M, Feinberg, H, Tucker, P.A, Shoham, G. | | Deposit date: | 1994-11-22 | | Release date: | 1996-08-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Carboxypeptidase A: native, zinc-removed and mercury-replaced forms.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1ARM
 
 | | CARBOXYPEPTIDASE A WITH ZN REPLACED BY HG | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COPPER (II) ION, HG-CARBOXYPEPTIDASE A=ALPHA= (COX), ... | | Authors: | Greenblatt, H.M, Feinberg, H, Tucker, P.A, Shoham, G. | | Deposit date: | 1994-11-22 | | Release date: | 1996-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Carboxypeptidase A: native, zinc-removed and mercury-replaced forms.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
2WB0
 
 | |
