1ABR
 
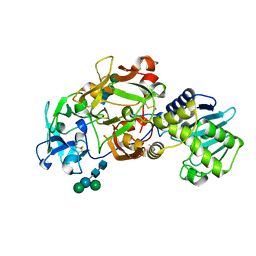 | | CRYSTAL STRUCTURE OF ABRIN-A | | Descriptor: | ABRIN-A, beta-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-L-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, beta-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Tahirov, T.H, Lu, T.-H, Liaw, Y.-C, Chu, S.-C, Lin, J.-Y. | | Deposit date: | 1994-11-11 | | Release date: | 1995-02-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of abrin-a at 2.14 A.
J.Mol.Biol., 250, 1995
|
|
1CPR
 
 | | ST. LOUIS CYTOCHROME C' FROM THE PURPLE PHOTOTROPIC BACTERIUM, RHODOBACTER CAPSULATUS | | Descriptor: | CYTOCHROME C', PROTOPORPHYRIN IX CONTAINING FE, ZINC ION | | Authors: | Tahirov, T.H, Misaki, S, Meyer, T.E, Cusanovich, M.A, Yasuoka, N. | | Deposit date: | 1996-04-09 | | Release date: | 1997-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of cytochrome c' from Rhodobacter capsulatus strain St Louis: an unusual molecular association induced by bridging Zn ions.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1GVD
 
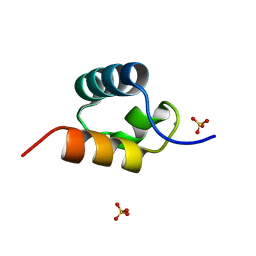 | | CRYSTAL STRUCTURE OF C-MYB R2 V103L MUTANT | | Descriptor: | AMMONIUM ION, MYB PROTO-ONCOGENE PROTEIN, SULFATE ION | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2002-02-08 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of C-Myb DNA-Binding Domain: Specific Na+ Binding and Correlation with NMR Structure
To be Published
|
|
1GTW
 
 | |
1GU4
 
 | |
1GU5
 
 | |
1H8A
 
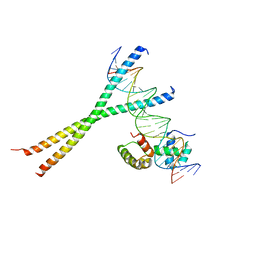 | | CRYSTAL STRUCTURE OF TERNARY PROTEIN-DNA COMPLEX3 | | Descriptor: | CAAT/ENHANCER BINDING PROTEIN BETA, DNA(5'-(*CP*CP*AP*GP*TP*CP*CP*GP*TP*TP*AP* AP*GP*GP*AP*TP*TP*GP*CP*GP*CP*CP*AP*CP*AP*T)-3'), DNA(5'-(*GP*AP*TP*GP*TP*GP*GP*CP*GP*CP*AP* AP*TP*CP*CP*TP*TP*AP*AP*CP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2001-01-31 | | Release date: | 2002-01-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Mechanism of C-Myb-C/Ebpbeta Cooperation from Separated Sites on a Promoter
Cell(Cambridge,Mass.), 108, 2002
|
|
1H89
 
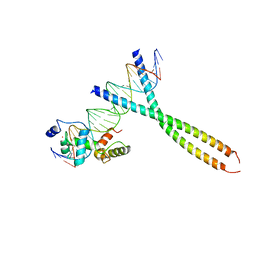 | | CRYSTAL STRUCTURE OF TERNARY PROTEIN-DNA COMPLEX2 | | Descriptor: | CAAT/ENHANCER BINDING PROTEIN BETA, DNA(5'-(*CP*CP*AP*GP*TP*CP*CP*GP*TP*TP*AP* AP*GP*GP*AP*TP*TP*GP*CP*GP*CP*CP*AP*CP*AP*T)-3'), DNA(5'-(*GP*AP*TP*GP*TP*GP*GP*CP*GP*CP*AP* AP*TP*CP*CP*TP*TP*AP*AP*CP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2001-01-30 | | Release date: | 2002-01-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Mechanism of C-Myb-C/Ebpbeta Cooperation from Separated Sites on a Promoter
Cell(Cambridge,Mass.), 108, 2002
|
|
1NBB
 
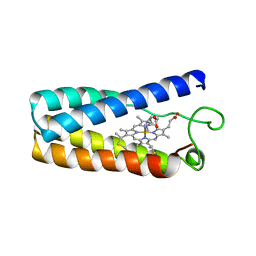 | | N-BUTYLISOCYANIDE BOUND RHODOBACTER CAPSULATUS CYTOCHROME C' | | Descriptor: | CYTOCHROME C', N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tahirov, T.H, Misaki, S, Meyer, T.E, Cusanovich, M.A, Higuchi, Y, Yasuoka, N. | | Deposit date: | 1996-03-18 | | Release date: | 1996-08-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Concerted movement of side chains in the haem vicinity observed on ligand binding in cytochrome c' from rhodobacter capsulatus.
Nat.Struct.Biol., 3, 1996
|
|
1H88
 
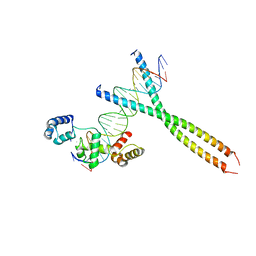 | | CRYSTAL STRUCTURE OF TERNARY PROTEIN-DNA COMPLEX1 | | Descriptor: | AMMONIUM ION, CCAAT/ENHANCER BINDING PROTEIN BETA, DNA(5'-(*CP*CP*AP*GP*TP*CP*CP*GP*TP*TP*AP* AP*GP*GP*AP*TP*TP*GP*CP*GP*CP*CP*AP*CP*AP*T)-3'), ... | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2001-01-29 | | Release date: | 2002-01-28 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of C-Myb-C/Ebpbeta Cooperation from Separated Sites on a Promoter
Cell(Cambridge,Mass.), 108, 2002
|
|
1CPQ
 
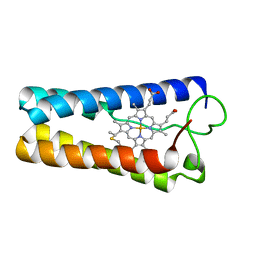 | | CYTOCHROME C' FROM RHODOPSEUDOMONAS CAPSULATA | | Descriptor: | CYTOCHROME C', PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tahirov, T.H, Misaki, S, Meyer, T.E, Cusanovich, M.A, Higuchi, Y, Yasuoka, N. | | Deposit date: | 1995-08-14 | | Release date: | 1996-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | High-resolution crystal structures of two polymorphs of cytochrome c' from the purple phototrophic bacterium rhodobacter capsulatus.
J.Mol.Biol., 259, 1996
|
|
1H38
 
 | | Structure of a T7 RNA polymerase elongation complex at 2.9A resolution | | Descriptor: | 5'-D(*GP*GP*GP*AP*AP*TP*CP*GP*AP*CP *AP*TP*CP*GP*CP*CP*GP*C)-3', 5'-D(*GP*TP*CP*GP*AP*TP*TP*CP*CP*CP)-3', 5'-R(*AP*AP*CP*UP*GP*CP*GP*GP*CP*GP *AP*U)-3', ... | | Authors: | Tahirov, T.H, Temyakov, D, Anikin, M, Patlan, V, McAllister, W.T, Vassylyev, D.G, Yokoyama, S. | | Deposit date: | 2002-08-24 | | Release date: | 2002-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a T7 RNA Polymerase Elongation Complex at 2.9 A Resolution
Nature, 420, 2002
|
|
1GV5
 
 | | CRYSTAL STRUCTURE OF C-MYB R2 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN, SODIUM ION | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2002-02-06 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of C-Myb DNA-Binding Domain: Specific Na+ Binding and Correlation with NMR Structure
To be Published
|
|
1HJB
 
 | |
1HJC
 
 | |
1IO4
 
 | |
3MI9
 
 | | Crystal structure of HIV-1 Tat complexed with human P-TEFb | | Descriptor: | Cell division protein kinase 9, Cyclin-T1, Protein Tat, ... | | Authors: | Tahirov, T.H, Babayeva, N.D, Varzavand, K, Cooper, J.J, Sedore, S.C, Price, D.H. | | Deposit date: | 2010-04-09 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of HIV-1 Tat complexed with human P-TEFb.
Nature, 465, 2010
|
|
3MIA
 
 | | Crystal structure of HIV-1 Tat complexed with ATP-bound human P-TEFb | | Descriptor: | Cell division protein kinase 9, Cyclin-T1, MAGNESIUM ION, ... | | Authors: | Tahirov, T.H, Babayeva, N.D, Varzavand, K, Cooper, J.J, Sedore, S.C, Price, D.H. | | Deposit date: | 2010-04-09 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of HIV-1 Tat complexed with human P-TEFb.
Nature, 465, 2010
|
|
1GUU
 
 | | CRYSTAL STRUCTURE OF C-MYB R1 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN, SODIUM ION | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2002-01-30 | | Release date: | 2003-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of C-Myb DNA-Binding Domain: Specific Na+ Binding and Correlation with NMR Structure
To be Published
|
|
1GV2
 
 | | CRYSTAL STRUCTURE OF C-MYB R2R3 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN, SODIUM ION | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2002-02-05 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of C-Myb DNA-Binding Domain: Specific Na+ Binding and Correlation with NMR Structure
To be Published
|
|
1ODJ
 
 | |
1ODI
 
 | |
1ODL
 
 | | PURINE NUCLEOSIDE PHOSPHORYLASE FROM THERMUS THERMOPHILUS | | Descriptor: | CHLORIDE ION, GLYCEROL, PURINE NUCLEOSIDE PHOSPHORYLASE, ... | | Authors: | Tahirov, T.H, Inagaki, E, Miyano, M. | | Deposit date: | 2003-02-19 | | Release date: | 2003-02-27 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Purine Nucleoside Phosphorylase from Thermus Thermophilus
J.Mol.Biol., 337, 2004
|
|
1ODK
 
 | |
1RCP
 
 | | CYTOCHROME C' | | Descriptor: | CYTOCHROME C', PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tahirov, T.H, Misaki, S, Meyer, T.E, Cusanovich, M.A, Higuchi, Y, Yasuoka, N. | | Deposit date: | 1995-08-23 | | Release date: | 1996-06-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution crystal structures of two polymorphs of cytochrome c' from the purple phototrophic bacterium rhodobacter capsulatus.
J.Mol.Biol., 259, 1996
|
|
