5RUB
 
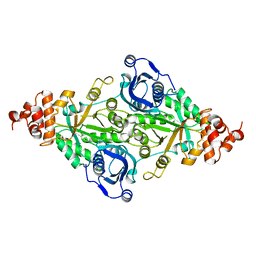 | | CRYSTALLOGRAPHIC REFINEMENT AND STRUCTURE OF RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE FROM RHODOSPIRILLUM RUBRUM AT 1.7 ANGSTROMS RESOLUTION | | Descriptor: | RUBISCO (RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE) | | Authors: | Schneider, G, Lindqvist, Y, Lundqvist, T. | | Deposit date: | 1990-05-29 | | Release date: | 1991-10-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic refinement and structure of ribulose-1,5-bisphosphate carboxylase from Rhodospirillum rubrum at 1.7 A resolution.
J.Mol.Biol., 211, 1990
|
|
6HNG
 
 | |
6HNE
 
 | |
4CSQ
 
 | |
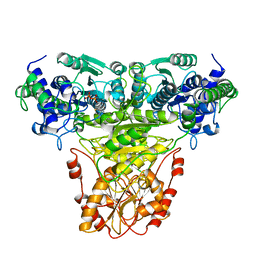
1TKB
 
 | |
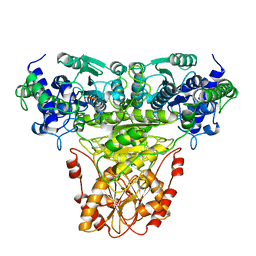
1TKA
 
 | |
1TKC
 
 | |
5MXL
 
 | |
5MXT
 
 | |
5OJH
 
 | | Crystal structure of the extramembrane domain of the cellulose biosynthetic protein BcsG from Salmonella typhimurium | | Descriptor: | CITRATE ANION, Cellulose biosynthesis protein BcsG, ZINC ION | | Authors: | Schneider, G, Vella, P, Lindqvist, Y, Schnell, R. | | Deposit date: | 2017-07-21 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and Functional Characterization of the BcsG Subunit of the Cellulose Synthase in Salmonella typhimurium.
J. Mol. Biol., 430, 2018
|
|
3EUL
 
 | | Structure of the signal receiver domain of the putative response regulator NarL from Mycobacterium tuberculosis | | Descriptor: | CHLORIDE ION, POSSIBLE NITRATE/NITRITE RESPONSE TRANSCRIPTIONAL REGULATORY PROTEIN NARL (DNA-binding response regulator, LuxR family) | | Authors: | Schneider, G, Schnell, R, Agren, D. | | Deposit date: | 2008-10-10 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 A structure of the signal receiver domain of the putative response regulator NarL from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
4IP2
 
 | | Putative Aromatic Acid Decarboxylase | | Descriptor: | 1,2-ETHANEDIOL, Aromatic Acid Decarboxylase, GLYCEROL, ... | | Authors: | Schneider, G, Brunner, K, Izumi, A, Jacewicz, A. | | Deposit date: | 2013-01-09 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into the UbiD protein family from the crystal structure of PA0254 from Pseudomonas aeruginosa.
Plos One, 8, 2013
|
|
3R8R
 
 | |
1RBA
 
 | |
2Q3D
 
 | |
2Q3C
 
 | |
2Q3B
 
 | |
6HNH
 
 | |
1F05
 
 | |
9RUB
 
 | | CRYSTAL STRUCTURE OF ACTIVATED RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE COMPLEXED WITH ITS SUBSTRATE, RIBULOSE-1,5-BISPHOSPHATE | | Descriptor: | FORMIC ACID, MAGNESIUM ION, RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE, ... | | Authors: | Lundqvist, T, Schneider, G. | | Deposit date: | 1990-11-28 | | Release date: | 1993-01-15 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of activated ribulose-1,5-bisphosphate carboxylase complexed with its substrate, ribulose-1,5-bisphosphate.
J.Biol.Chem., 266, 1991
|
|
3KOF
 
 | | Crystal structure of the double mutant F178Y/R181E of E.coli transaldolase B | | Descriptor: | SULFATE ION, Transaldolase B | | Authors: | Schneider, S, Gutierrez, M, Sandalova, T, Schneider, G, Clapes, P, Sprenger, G.A, Samland, A.K. | | Deposit date: | 2009-11-13 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redesigning the Active Site of Transaldolase TalB from Escherichia coli: New Variants with Improved Affinity towards Nonphosphorylated Substrates.
Chembiochem, 11, 2010
|
|
3KG1
 
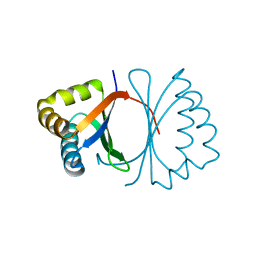 | | Crystal structure of SnoaB, a cofactor-independent oxygenase from Streptomyces nogalater, mutant N63A | | Descriptor: | CHLORIDE ION, SnoaB | | Authors: | Koskiniemi, H, Grocholski, T, Lindqvist, Y, Mantsala, P, Niemi, J, Schneider, G. | | Deposit date: | 2009-10-28 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the cofactor-independent monooxygenase SnoaB from Streptomyces nogalater: implications for the reaction mechanism
Biochemistry, 49, 2010
|
|
3KG0
 
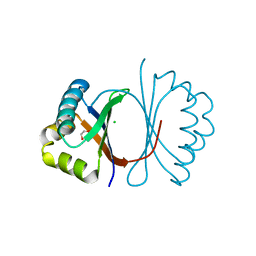 | | Crystal structure of SnoaB, a cofactor-independent oxygenase from Streptomyces nogalater, determined to 1.7 resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SnoaB | | Authors: | Koskiniemi, H, Grocholski, T, Lindqvist, Y, Mantsala, P, Niemi, J, Schneider, G. | | Deposit date: | 2009-10-28 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the cofactor-independent monooxygenase SnoaB from Streptomyces nogalater: implications for the reaction mechanism
Biochemistry, 49, 2010
|
|
6EWY
 
 | | RipA Peptidoglycan hydrolase (Rv1477, Mycobacterium tuberculosis) N-terminal domain | | Descriptor: | Peptidoglycan endopeptidase RipA | | Authors: | Schnell, R, Steiner, E.M, Schneider, G, Guy, J, Bourenkov, G. | | Deposit date: | 2017-11-07 | | Release date: | 2018-05-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of the N-terminal module of the cell wall hydrolase RipA and its role in regulating catalytic activity.
Proteins, 86, 2018
|
|
4LJ1
 
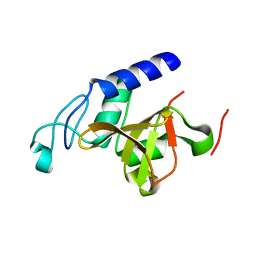 | | RipD (Rv1566c) from Mycobacterium tuberculosis: a non-catalytic NlpC/p60 domain protein with two penta-peptide repeat units (PVQQA-PVQPA) | | Descriptor: | Invasion-associated protein | | Authors: | Both, D, Steiner, E.M, Linder, D.C, Schnell, R, Schneider, G. | | Deposit date: | 2013-07-04 | | Release date: | 2013-11-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | RipD (Rv1566c) from Mycobacterium tuberculosis: adaptation of an NlpC/p60 domain to a non-catalytic peptidoglycan-binding function.
Biochem.J., 457, 2014
|
|
