1MJC
 
 | |
1CSQ
 
 | |
1CSP
 
 | |
1N2C
 
 | | NITROGENASE COMPLEX FROM AZOTOBACTER VINELANDII STABILIZED BY ADP-TETRAFLUOROALUMINATE | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Schindelin, H, Kisker, C, Rees, D.C. | | Deposit date: | 1997-05-02 | | Release date: | 1997-11-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of ADP x AIF4(-)-stabilized nitrogenase complex and its implications for signal transduction.
Nature, 387, 1997
|
|
1SDR
 
 | | CRYSTAL STRUCTURE OF AN RNA DODECAMER CONTAINING THE ESCHERICHIA COLI SHINE-DALGARNO SEQUENCE | | Descriptor: | RNA (5'-R(*AP*UP*CP*AP*CP*CP*UP*CP*CP*UP*UP*A)-3'), RNA (5'-R(*UP*AP*AP*GP*GP*AP*GP*GP*UP*GP*AP*U)-3') | | Authors: | Schindelin, H, Zhang, M, Bald, R, Fuerste, J.-P, Erdmann, V.A, Heinemann, U. | | Deposit date: | 1994-12-11 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of an RNA dodecamer containing the Escherichia coli Shine-Dalgarno sequence.
J.Mol.Biol., 249, 1995
|
|
2B5E
 
 | | Crystal Structure of Yeast Protein Disulfide Isomerase | | Descriptor: | BARIUM ION, GLYCEROL, Protein disulfide-isomerase | | Authors: | Schindelin, H, Tian, G. | | Deposit date: | 2005-09-28 | | Release date: | 2006-01-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of yeast protein disulfide isomerase suggests cooperativity between its active sites.
Cell(Cambridge,Mass.), 124, 2006
|
|
7PKJ
 
 | | Streptococcus pyogenes apo GapN | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, Putative NADP-dependent glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Schindelin, H, Albert, L. | | Deposit date: | 2021-08-25 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | The Non-phosphorylating Glyceraldehyde-3-Phosphate Dehydrogenase GapN Is a Potential New Drug Target in Streptococcus pyogenes.
Front Microbiol, 13, 2022
|
|
7PKC
 
 | | Streptococcus pyogenes Apo-GapN C284S variant | | Descriptor: | GLYCEROL, Putative NADP-dependent glyceraldehyde-3-phosphate dehydrogenase, SULFATE ION | | Authors: | Schindelin, H, Albert, L. | | Deposit date: | 2021-08-25 | | Release date: | 2022-03-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Non-phosphorylating Glyceraldehyde-3-Phosphate Dehydrogenase GapN Is a Potential New Drug Target in Streptococcus pyogenes.
Front Microbiol, 13, 2022
|
|
1EKS
 
 | | ASP128ALA VARIANT OF MOAC PROTEIN FROM E. COLI | | Descriptor: | L(+)-TARTARIC ACID, MOLYBDENUM COFACTOR BIOSYNTHESIS PROTEIN C | | Authors: | Schindelin, H, Liu, M.T.W, Wuebbens, M.M, Rajagopalan, K.V. | | Deposit date: | 2000-03-09 | | Release date: | 2000-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into molybdenum cofactor deficiency provided by the crystal structure of the molybdenum cofactor biosynthesis protein MoaC.
Structure Fold.Des., 8, 2000
|
|
1EKR
 
 | | MOAC PROTEIN FROM E. COLI | | Descriptor: | MOLYBDENUM COFACTOR BIOSYNTHESIS PROTEIN C | | Authors: | Schindelin, H, Liu, M.T.W, Wuebbens, M.M, Rajagopalan, K.V. | | Deposit date: | 2000-03-09 | | Release date: | 2000-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into molybdenum cofactor deficiency provided by the crystal structure of the molybdenum cofactor biosynthesis protein MoaC.
Structure Fold.Des., 8, 2000
|
|
4V98
 
 | | The 8S snRNP Assembly Intermediate | | Descriptor: | CG10419, Icln, LD23602p, ... | | Authors: | Grimm, C, Pelz, J.P, Schindelin, H, Diederichs, K, Kuper, J, Kisker, C. | | Deposit date: | 2012-05-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Assembly Chaperone- Mediated snRNP Formation.
Mol.Cell, 49, 2013
|
|
5AES
 
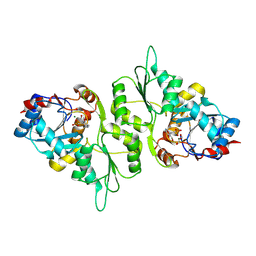 | | Crystal Structure of murine Chronophin (Pyridoxal Phosphate Phosphatase) in Complex with a PNP-derived Inhibitor | | Descriptor: | GLYCEROL, MAGNESIUM ION, PYRIDOXAL PHOSPHATE PHOSPHATASE, ... | | Authors: | Knobloch, G, Jabari, N, Koehn, M, Gohla, A, Schindelin, H. | | Deposit date: | 2015-01-09 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Synthesis of Hydrolysis-Resistant Pyridoxal 5'-Phosphate Analogs and Their Biochemical and X-Ray Crystallographic Characterization with the Pyridoxal Phosphatase Chronophin.
Bioorg.Med.Chem., 23, 2015
|
|
5L6H
 
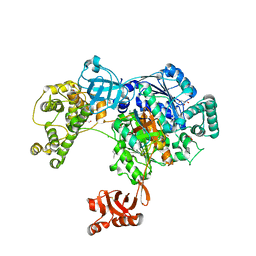 | | Uba1 in complex with Ub-ABPA3 covalent adduct | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2016-05-30 | | Release date: | 2017-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dissecting the Specificity of Adenosyl Sulfamate Inhibitors Targeting the Ubiquitin-Activating Enzyme.
Structure, 25, 2017
|
|
5L6I
 
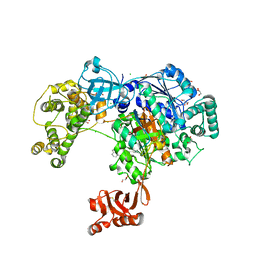 | | Uba1 in complex with Ub-MLN4924 covalent adduct | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2016-05-30 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Dissecting the Specificity of Adenosyl Sulfamate Inhibitors Targeting the Ubiquitin-Activating Enzyme.
Structure, 25, 2017
|
|
5L6J
 
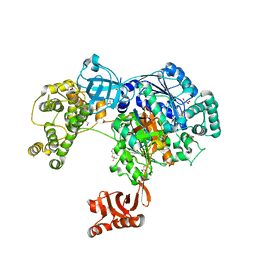 | | Uba1 in complex with Ub-MLN7243 covalent adduct | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2016-05-30 | | Release date: | 2017-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Dissecting the Specificity of Adenosyl Sulfamate Inhibitors Targeting the Ubiquitin-Activating Enzyme.
Structure, 25, 2017
|
|
7ZH9
 
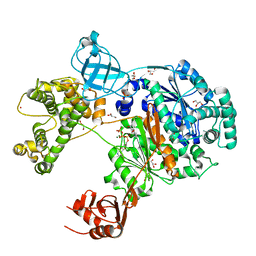 | | Uba1 in complex with ATP | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2022-04-05 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structures of UBA6 explain its dual specificity for ubiquitin and FAT10.
Nat Commun, 13, 2022
|
|
7ZTL
 
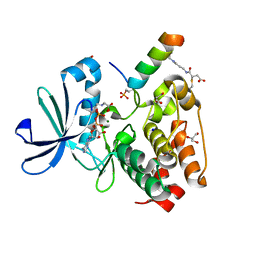 | |
8A1I
 
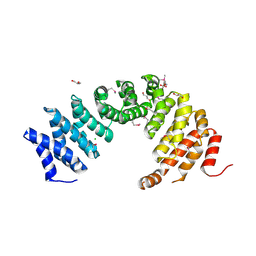 | |
6ZQH
 
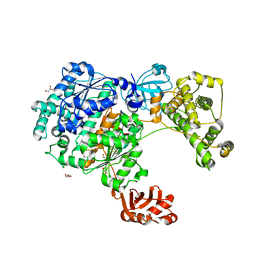 | | Yeast Uba1 in complex with ubiquitin | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, TETRAETHYLENE GLYCOL, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.032 Å) | | Cite: | Development of ADPribosyl Ubiquitin Analogues to Study Enzymes Involved in Legionella Infection.
Chemistry, 27, 2021
|
|
4F9Z
 
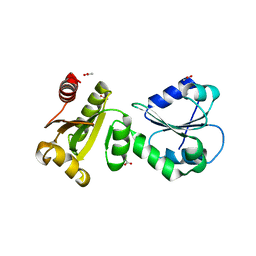 | | Crystal Structure of human ERp27 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ACETATE ION, ... | | Authors: | Kober, F.X, Koelmel, W, Kuper, J, Schindelin, H. | | Deposit date: | 2012-05-21 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of the Protein-Disulfide Isomerase Family Member ERp27 Provides Insights into Its Substrate Binding Capabilities.
J.Biol.Chem., 288, 2013
|
|
3BII
 
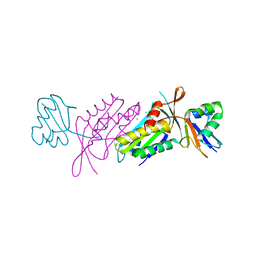 | | Crystal Structure of Activated MPT Synthase | | Descriptor: | CHLORIDE ION, Molybdopterin-converting factor subunit 1, Molybdopterin-converting factor subunit 2 | | Authors: | Daniels, J.N, Schindelin, H. | | Deposit date: | 2007-11-30 | | Release date: | 2008-02-19 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a molybdopterin synthase-precursor Z complex: insight into its sulfur transfer mechanism and its role in molybdenum cofactor deficiency.
Biochemistry, 47, 2008
|
|
2AF4
 
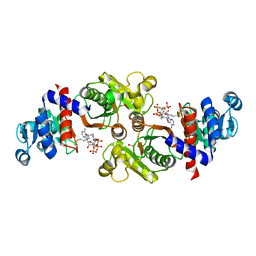 | | Phosphotransacetylase from Methanosarcina thermophila co-crystallized with coenzyme A | | Descriptor: | COENZYME A, Phosphate acetyltransferase | | Authors: | Lawrence, S.H, Luther, K.B, Ferry, J.G, Schindelin, H. | | Deposit date: | 2005-07-25 | | Release date: | 2006-01-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Structural and functional studies suggest a catalytic mechanism for the phosphotransacetylase from Methanosarcina thermophila.
J.Bacteriol., 188, 2006
|
|
2AF3
 
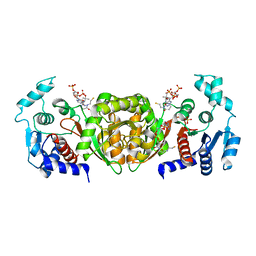 | | Phosphotransacetylase from Methanosarcina thermophila soaked with Coenzyme A | | Descriptor: | COENZYME A, Phosphate acetyltransferase, SULFATE ION | | Authors: | Lawrence, S.H, Luther, K.B, Ferry, J.G, Schindelin, H. | | Deposit date: | 2005-07-25 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional studies suggest a catalytic mechanism for the phosphotransacetylase from Methanosarcina thermophila.
J.Bacteriol., 188, 2006
|
|
3GAE
 
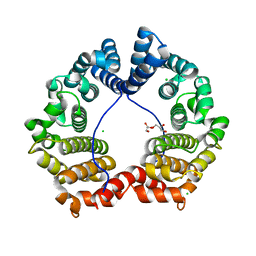 | | Crystal Structure of PUL | | Descriptor: | CHLORIDE ION, GLYCEROL, Protein DOA1 | | Authors: | Zhao, G, Schindelin, H, Lennarz, W.J. | | Deposit date: | 2009-02-17 | | Release date: | 2009-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An Armadillo motif in Ufd3 interacts with Cdc48 and is involved in ubiquitin homeostasis and protein degradation
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3IPO
 
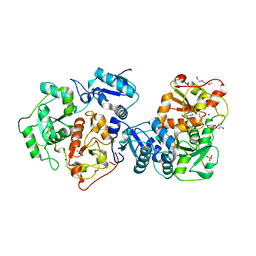 | | Crystal structure of YnjE | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, GLYCEROL, ... | | Authors: | Haenzelmann, P, Kuper, J, Schindelin, H. | | Deposit date: | 2009-08-18 | | Release date: | 2009-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of YnjE from Escherichia coli, a sulfurtransferase with three rhodanese domains.
Protein Sci., 18, 2009
|
|
