7VFQ
 
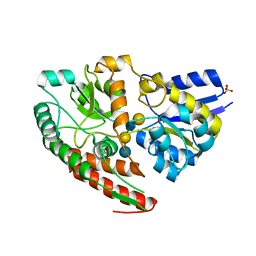 | | Wild type GltA from Bifidobacterium infantis JCM 1222 complexed with lacto-N-tetraose | | Descriptor: | Extracellular solute-binding protein, family 1, SULFATE ION, ... | | Authors: | Sato, M, Sakanaka, M, Katayama, T, Fushinobu, S. | | Deposit date: | 2021-09-13 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Short stretch sequence of a milk oligosaccharide transporter represents the phenotypic selection trajectory in infant gut microbiome
To Be Published
|
|
6HUS
 
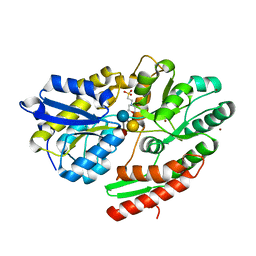 | | 2'-fucosyllactose and 3-fucosyllactose binding protein from Bifidobacterium longum infantis, bound with 3-fucosyllactose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ABC transporter substrate-binding protein, ZINC ION, ... | | Authors: | Ejby, M, Abou Hachem, M, Lo Leggio, L, Takane, K, Sakanaka, M. | | Deposit date: | 2018-10-09 | | Release date: | 2019-09-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.409 Å) | | Cite: | Evolutionary adaptation in fucosyllactose uptake systems supports bifidobacteria-infant symbiosis.
Sci Adv, 5, 2019
|
|
6HUR
 
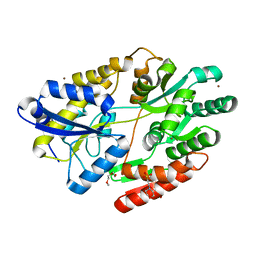 | | 2'-fucosyllactose and 3-fucosyllactose binding protein from Bifidobacterium longum infantis, bound with 2'-fucosyllactose | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ABC transporter substrate-binding protein, ... | | Authors: | Ejby, M, Abou Hachem, M, Lo Leggio, L, Katayama, T, Sakanaka, M. | | Deposit date: | 2018-10-09 | | Release date: | 2019-09-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.297 Å) | | Cite: | Evolutionary adaptation in fucosyllactose uptake systems supports bifidobacteria-infant symbiosis.
Sci Adv, 5, 2019
|
|
7VFR
 
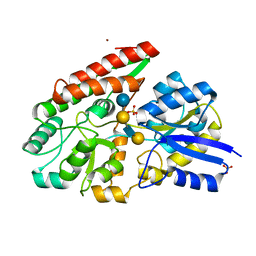 | | GltA N83K mutant from Bifidobacterium infantis JCM 1222 complexed with lacto-N-tetraose | | Descriptor: | Extracellular solute-binding protein, family 1, SULFATE ION, ... | | Authors: | Sato, M, Sakanaka, M, Katayama, T, Fushinobu, S. | | Deposit date: | 2021-09-13 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Short stretch sequence of a milk oligosaccharide transporter represents the phenotypic selection trajectory in infant gut microbiome
To Be Published
|
|
8IBS
 
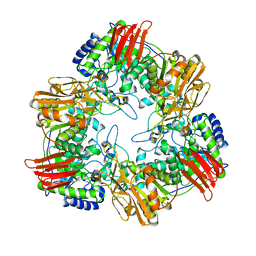 | | Crystal structure of GH42 beta-galactosidase BiBga42A from Bifidobacterium longum subspecies infantis E160A/E318A mutant in complex with galactose | | Descriptor: | Beta-galactosidase, alpha-D-galactopyranose | | Authors: | Hidaka, M, Fushinobu, S, Gotoh, A, Katayama, T. | | Deposit date: | 2023-02-10 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate recognition mode of a glycoside hydrolase family 42 beta-galactosidase from Bifidobacterium longum subspecies infantis ( Bi Bga42A) revealed by crystallographic and mutational analyses.
Microbiome Res Rep, 2, 2023
|
|
8IBT
 
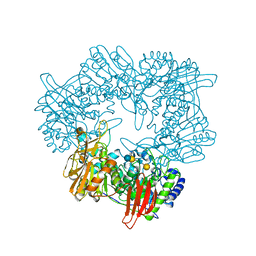 | | Crystal structure of GH42 beta-galactosidase BiBga42A from Bifidobacterium longum subspecies infantis E318S mutant in complex with lacto-N-tetraose | | Descriptor: | Beta-galactosidase, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Hidaka, M, Fushinobu, S, Gotoh, A, Katayama, T. | | Deposit date: | 2023-02-10 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate recognition mode of a glycoside hydrolase family 42 beta-galactosidase from Bifidobacterium longum subspecies infantis ( Bi Bga42A) revealed by crystallographic and mutational analyses.
Microbiome Res Rep, 2, 2023
|
|
8IBR
 
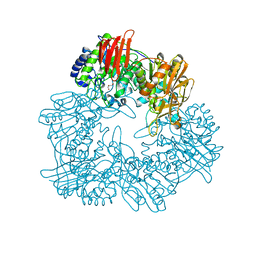 | | Crystal structure of GH42 beta-galactosidase BiBga42A from Bifidobacterium longum subspecies infantis in complex with glycerol | | Descriptor: | Beta-galactosidase, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Hidaka, M, Fushinobu, S, Gotoh, A, Katayama, T. | | Deposit date: | 2023-02-10 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate recognition mode of a glycoside hydrolase family 42 beta-galactosidase from Bifidobacterium longum subspecies infantis ( Bi Bga42A) revealed by crystallographic and mutational analyses.
Microbiome Res Rep, 2, 2023
|
|
5GQC
 
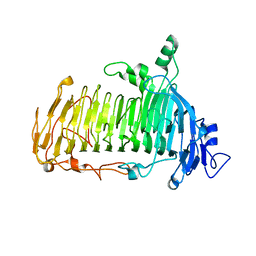 | | Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form | | Descriptor: | CALCIUM ION, Lacto-N-biosidase, SODIUM ION | | Authors: | Yamada, C, Arakawa, T, Katayama, T, Fushinobu, S. | | Deposit date: | 2016-08-07 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Molecular Insight into Evolution of Symbiosis between Breast-Fed Infants and a Member of the Human Gut Microbiome Bifidobacterium longum
Cell Chem Biol, 24, 2017
|
|
5GQG
 
 | | Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex | | Descriptor: | CALCIUM ION, Lacto-N-biosidase, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose | | Authors: | Yamada, C, Arakawa, T, Katayama, T, Fushinobu, S. | | Deposit date: | 2016-08-07 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Insight into Evolution of Symbiosis between Breast-Fed Infants and a Member of the Human Gut Microbiome Bifidobacterium longum
Cell Chem Biol, 24, 2017
|
|
5GQF
 
 | | Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex | | Descriptor: | CALCIUM ION, Lacto-N-biosidase, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yamada, C, Arakawa, T, Katayama, T, Fushinobu, S. | | Deposit date: | 2016-08-07 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Molecular Insight into Evolution of Symbiosis between Breast-Fed Infants and a Member of the Human Gut Microbiome Bifidobacterium longum
Cell Chem Biol, 24, 2017
|
|
6KQT
 
 | | Crystal Structure of GH136 lacto-N-biosidase from Eubacterium ramulus - native protein | | Descriptor: | SODIUM ION, TRIETHYLENE GLYCOL, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamada, C, Arakawa, T, Pichler, M.J, Abou Hachem, M, Fushinobu, S. | | Deposit date: | 2019-08-18 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Butyrate producing colonic Clostridiales metabolise human milk oligosaccharides and cross feed on mucin via conserved pathways.
Nat Commun, 11, 2020
|
|
6KQS
 
 | | Crystal Structure of GH136 lacto-N-biosidase from Eubacterium ramulus - selenomethionine derivative | | Descriptor: | GLYCEROL, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, lacto-N-biosidase | | Authors: | Yamada, C, Arakawa, T, Pichler, M.J, Abou Hachem, M, Fushinobu, S. | | Deposit date: | 2019-08-18 | | Release date: | 2020-06-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Butyrate producing colonic Clostridiales metabolise human milk oligosaccharides and cross feed on mucin via conserved pathways.
Nat Commun, 11, 2020
|
|
7WDT
 
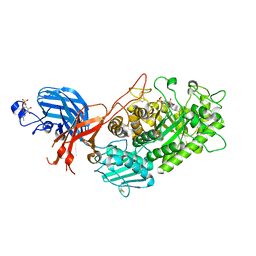 | | 6-sulfo-beta-D-N-acetylglucosaminidase from Bifidobacterium bifidum in complex with GlcNAc-6S | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, ... | | Authors: | Yamada, C, Kashima, T, Fushinobu, S, Katoh, T, Katayama, T. | | Deposit date: | 2021-12-22 | | Release date: | 2022-12-28 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A bacterial sulfoglycosidase highlights mucin O-glycan breakdown in the gut ecosystem.
Nat.Chem.Biol., 19, 2023
|
|
7WDU
 
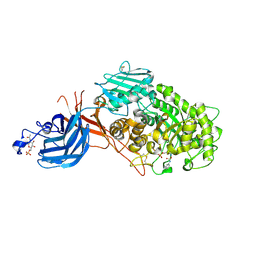 | | 6-sulfo-beta-D-N-acetylglucosaminidase from Bifidobacterium bifidum in complex with PUGNAc-6S | | Descriptor: | Beta-N-acetylhexosaminidase, CALCIUM ION, [[(3R,4R,5S,6R)-3-acetamido-4,5-bis(oxidanyl)-6-(sulfooxymethyl)oxan-2-ylidene]amino] N-phenylcarbamate | | Authors: | Kashima, T, Yamada, C, Fushinobu, S, Katoh, T, Katayama, T. | | Deposit date: | 2021-12-22 | | Release date: | 2022-12-28 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | A bacterial sulfoglycosidase highlights mucin O-glycan breakdown in the gut ecosystem.
Nat.Chem.Biol., 19, 2023
|
|
