4D0A
 
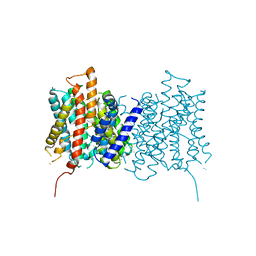 | |
5NL2
 
 | | cryo-EM structure of the mTMEM16A ion channel at 6.6 A resolution. | | Descriptor: | Anoctamin-1 | | Authors: | Paulino, C, Neldner, Y, Lam, K.M, Kalienkova, V, Brunner, J.D, Schenck, S, Dutzler, R. | | Deposit date: | 2017-04-03 | | Release date: | 2017-06-07 | | Last modified: | 2018-02-28 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural basis for anion conduction in the calcium-activated chloride channel TMEM16A.
Elife, 6, 2017
|
|
5OYB
 
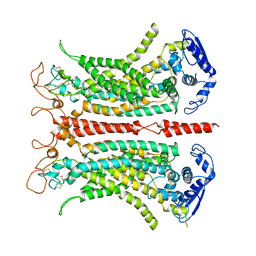 | | Structure of calcium-bound mTMEM16A chloride channel at 3.75 A resolution | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Paulino, C, Kalienkova, V, Lam, K.M, Neldner, Y, Dutzler, R. | | Deposit date: | 2017-09-08 | | Release date: | 2017-12-20 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Activation mechanism of the calcium-activated chloride channel TMEM16A revealed by cryo-EM.
Nature, 552, 2017
|
|
5OYG
 
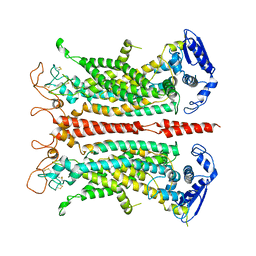 | | Structure of calcium-free mTMEM16A chloride channel at 4.06 A resolution | | Descriptor: | Anoctamin-1 | | Authors: | Paulino, C, Kalienkova, V, Lam, K.M, Neldner, Y, Dutzler, R. | | Deposit date: | 2017-09-08 | | Release date: | 2017-12-20 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Activation mechanism of the calcium-activated chloride channel TMEM16A revealed by cryo-EM.
Nature, 552, 2017
|
|
4CZB
 
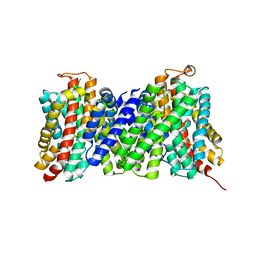 | | Structure of the sodium proton antiporter MjNhaP1 from Methanocaldococcus jannaschii at pH 8. | | Descriptor: | NA(+)/H(+) ANTIPORTER 1, POTASSIUM ION, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Woehlert, D, Paulino, C, Kapotova, E, Kuhlbrandt, W, Yildiz, O. | | Deposit date: | 2014-04-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | 3D Em Map of the Sodium Proton Antiporter Mjnhap1 from Methanocaldococcus Jannaschii
Elife, 3, 2014
|
|
8BMP
 
 | | Cryo-EM structure of the folate-specific ECF transporter complex in MSP2N2 lipid nanodiscs bound to ATP and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Energy-coupling factor transporter ATP-binding protein EcfA1, ... | | Authors: | Thangaratnarajah, C, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2022-11-10 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Expulsion mechanism of the substrate-translocating subunit in ECF transporters.
Nat Commun, 14, 2023
|
|
8BMS
 
 | | Cryo-EM structure of the mutant solitary ECF module 2EQ in MSP2N2 lipid nanodiscs in the ATPase closed and ATP-bound conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Energy-coupling factor transporter ATP-binding protein EcfA1, Energy-coupling factor transporter ATP-binding protein EcfA2, ... | | Authors: | Thangaratnarajah, C, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2022-11-10 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Expulsion mechanism of the substrate-translocating subunit in ECF transporters.
Nat Commun, 14, 2023
|
|
8BMR
 
 | | Cryo-EM structure of the wild-type solitary ECF module in MSP2N2 lipid nanodiscs in the ATPase open and nucleotide-free conformation | | Descriptor: | Energy-coupling factor transporter ATP-binding protein EcfA1, Energy-coupling factor transporter ATP-binding protein EcfA2, Energy-coupling factor transporter transmembrane protein EcfT | | Authors: | Thangaratnarajah, C, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2022-11-10 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Expulsion mechanism of the substrate-translocating subunit in ECF transporters.
Nat Commun, 14, 2023
|
|
8BMQ
 
 | | Cryo-EM structure of the folate-specific ECF transporter complex in MSP2N2 lipid nanodiscs bound to AMP-PNP | | Descriptor: | Energy-coupling factor transporter ATP-binding protein EcfA1, Energy-coupling factor transporter ATP-binding protein EcfA2, Energy-coupling factor transporter transmembrane protein EcfT, ... | | Authors: | Thangaratnarajah, C, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2022-11-10 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Expulsion mechanism of the substrate-translocating subunit in ECF transporters.
Nat Commun, 14, 2023
|
|
6HRA
 
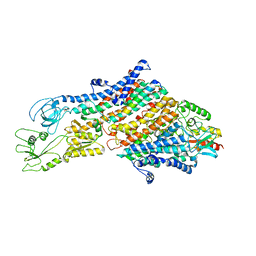 | | Cryo-EM structure of the KdpFABC complex in an E1 outward-facing state (state 1) | | Descriptor: | POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, Potassium-transporting ATPase KdpC subunit, ... | | Authors: | Stock, C, Hielkema, L, Tascon, I, Wunnicke, D, Oostergetel, G.T, Azkargorta, M, Paulino, C, Haenelt, I. | | Deposit date: | 2018-09-26 | | Release date: | 2018-12-05 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of KdpFABC suggest a K+transport mechanism via two inter-subunit half-channels.
Nat Commun, 9, 2018
|
|
6HRB
 
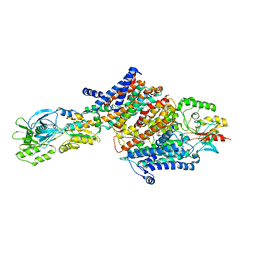 | | Cryo-EM structure of the KdpFABC complex in an E2 inward-facing state (state 2) | | Descriptor: | POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, Potassium-transporting ATPase KdpC subunit, ... | | Authors: | Stock, C, Hielkema, L, Tascon, I, Wunnicke, D, Oostergetel, G.T, Azkargorta, M, Paulino, C, Haenelt, I. | | Deposit date: | 2018-09-26 | | Release date: | 2018-12-05 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of KdpFABC suggest a K+transport mechanism via two inter-subunit half-channels.
Nat Commun, 9, 2018
|
|
7NNP
 
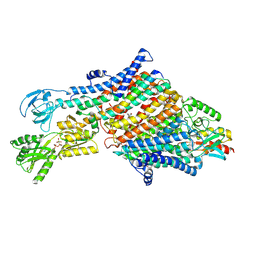 | | Rb-loaded cryo-EM structure of the E1-ATP KdpFABC complex. | | Descriptor: | CARDIOLIPIN, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Silberberg, J.M, Corey, R.A, Hielkema, L, Stock, C, Stansfeld, P.J, Paulino, C, Haenelt, I. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-28 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Deciphering ion transport and ATPase coupling in the intersubunit tunnel of KdpFABC.
Nat Commun, 12, 2021
|
|
7NNL
 
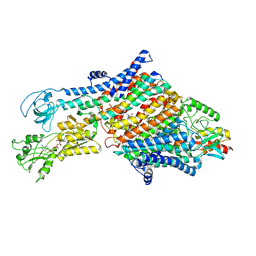 | | Cryo-EM structure of the KdpFABC complex in an E1-ATP conformation loaded with K+ | | Descriptor: | CARDIOLIPIN, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Silberberg, J.M, Corey, R.A, Hielkema, L, Stock, C, Stansfeld, P.J, Paulino, C, Haenelt, I. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-28 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Deciphering ion transport and ATPase coupling in the intersubunit tunnel of KdpFABC.
Nat Commun, 12, 2021
|
|
7BCS
 
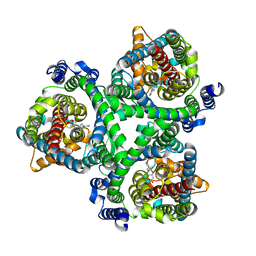 | | ASCT2 in the presence of the inhibitor Lc-BPE (position "down") in the outward-open conformation. | | Descriptor: | (2~{S},4~{S})-4-(4-phenylphenyl)carbonyloxypyrrolidine-2-carboxylic acid, Neutral amino acid transporter B(0) | | Authors: | Garibsingh, R.A, Ndaru, E, Garaeva, A.A, Shi, Y, Zielewicz, L, Bonomi, M, Slotboom, D.J, Paulino, C, Grewer, C, Schlessinger, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Rational design of ASCT2 inhibitors using an integrated experimental-computational approach.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6GCT
 
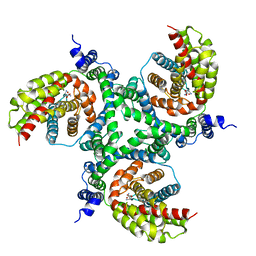 | | cryo-EM structure of the human neutral amino acid transporter ASCT2 | | Descriptor: | GLUTAMINE, Neutral amino acid transporter B(0) | | Authors: | Garaeva, A.A, Oostergetel, G.T, Gati, C, Guskov, A, Paulino, C, Slotboom, D.J. | | Deposit date: | 2018-04-19 | | Release date: | 2018-06-13 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Cryo-EM structure of the human neutral amino acid transporter ASCT2.
Nat. Struct. Mol. Biol., 25, 2018
|
|
8ON7
 
 | | FMRFa-bound Malacoceros FaNaC1 in lipid nanodiscs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FMRFamide, ... | | Authors: | Kalienkova, V, Dandamudi, M, Paulino, C, Lynagh, T. | | Deposit date: | 2023-04-01 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural basis for excitatory neuropeptide signaling.
Nat.Struct.Mol.Biol., 2024
|
|
8ONA
 
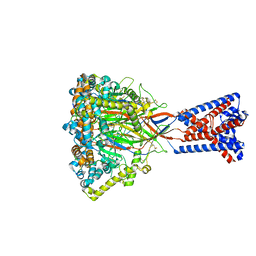 | | FMRFa-bound Malacoceros FaNaC1 in lipid nanodiscs in presence of diminazene | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FMRFamide, neuropeptide, ... | | Authors: | Kalienkova, V, Dandamudi, M, Paulino, C, Lynagh, T. | | Deposit date: | 2023-04-01 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for excitatory neuropeptide signaling.
Nat.Struct.Mol.Biol., 2024
|
|
8ON9
 
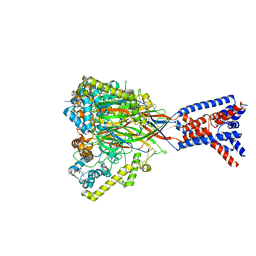 | | ASSFVRIa-bound Malacoceros FaNaC1 in lipid nanodiscs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ASSFVRIamide, ... | | Authors: | Kalienkova, V, Dandamudi, M, Paulino, C, Lynagh, T. | | Deposit date: | 2023-04-01 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis for excitatory neuropeptide signaling.
Nat.Struct.Mol.Biol., 2024
|
|
8ON8
 
 | | Apo Malacoceros FaNaC1 in lipid nanodiscs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FMRFamide-gated sodium channel 1 | | Authors: | Kalienkova, V, Dandamudi, M, Paulino, C, Lynagh, T. | | Deposit date: | 2023-04-01 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for excitatory neuropeptide signaling.
Nat.Struct.Mol.Biol., 2024
|
|
8PD8
 
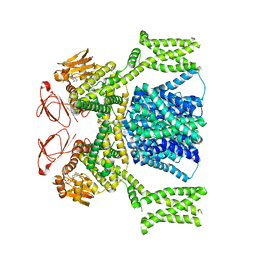 | | cAMP-bound SpSLC9C1 in lipid nanodiscs, dimer | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Sperm-specific sodium proton exchanger | | Authors: | Kalienkova, V, Peter, M, Rheinberger, J, Paulino, C. | | Deposit date: | 2023-06-12 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of a sperm-specific solute carrier gated by voltage and cAMP.
Nature, 623, 2023
|
|
8PD9
 
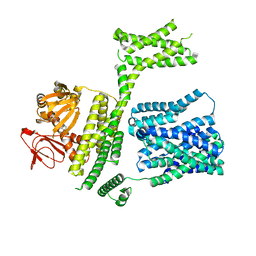 | | cAMP-bound SpSLC9C1 in lipid nanodiscs, protomer state 1 | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Sperm-specific sodium proton exchanger | | Authors: | Kalienkova, V, Peter, M, Rheinberger, J, Paulino, C. | | Deposit date: | 2023-06-12 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of a sperm-specific solute carrier gated by voltage and cAMP.
Nature, 623, 2023
|
|
8PD2
 
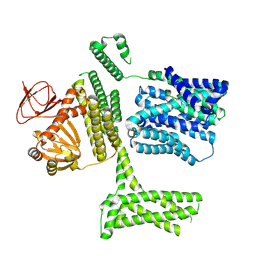 | | Ligand-free SpSLC9C1 in lipid nanodiscs, protomer state 1 | | Descriptor: | Sperm-specific sodium proton exchanger | | Authors: | Kalienkova, V, Peter, M, Rheinberger, J, Paulino, C. | | Deposit date: | 2023-06-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structures of a sperm-specific solute carrier gated by voltage and cAMP.
Nature, 623, 2023
|
|
8PD5
 
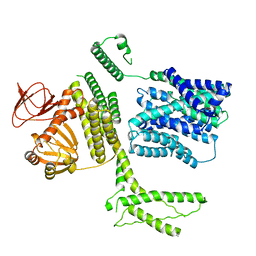 | | Ligand-free SpSLC9C1 in lipid nanodiscs, protomer state 3 | | Descriptor: | Sperm-specific sodium proton exchanger | | Authors: | Kalienkova, V, Peter, M, Rheinberger, J, Paulino, C. | | Deposit date: | 2023-06-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of a sperm-specific solute carrier gated by voltage and cAMP.
Nature, 623, 2023
|
|
8PDU
 
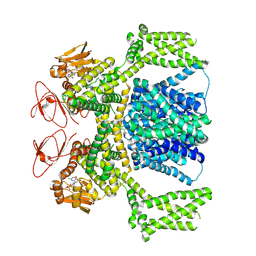 | | cGMP-bound SpSLC9C1 in lipid nanodiscs, dimer | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, Sperm-specific sodium proton exchanger | | Authors: | Kalienkova, V, Peter, M, Rheinberger, J, Paulino, C. | | Deposit date: | 2023-06-12 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structures of a sperm-specific solute carrier gated by voltage and cAMP.
Nature, 623, 2023
|
|
8PD7
 
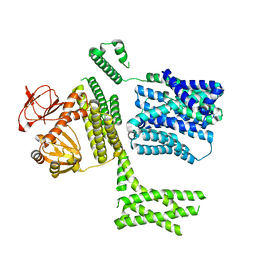 | | Ligand-free SpSLC9C1 in lipid nanodiscs, protomer state 4 | | Descriptor: | Sperm-specific sodium proton exchanger | | Authors: | Kalienkova, V, Peter, M, Rheinberger, J, Paulino, C. | | Deposit date: | 2023-06-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of a sperm-specific solute carrier gated by voltage and cAMP.
Nature, 623, 2023
|
|
