1CEK
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE MEMBRANE-EMBEDDED M2 CHANNEL-LINING SEGMENT FROM THE NICOTINIC ACETYLCHOLINE RECEPTOR BY SOLID-STATE NMR SPECTROSCOPY | | Descriptor: | PROTEIN (ACETYLCHOLINE RECEPTOR M2) | | Authors: | Marassi, F.M, Gesell, J.J, Kim, Y, Valente, A.P, Oblatt-Montal, M, Montal, M, Opella, S.J. | | Deposit date: | 1999-03-09 | | Release date: | 1999-03-11 | | Last modified: | 2023-12-27 | | Method: | SOLID-STATE NMR | | Cite: | Structures of the M2 channel-lining segments from nicotinic acetylcholine and NMDA receptors by NMR spectroscopy.
Nat.Struct.Biol., 6, 1999
|
|
1EQ8
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE PENTAMERIC HELICAL BUNDLE OF THE ACETYLCHOLINE RECEPTOR M2 TRANSMEMBRANE SEGMENT | | Descriptor: | ACETYLCHOLINE RECEPTOR PROTEIN, HYDROXIDE ION | | Authors: | Marassi, F.M, Gesell, J.J, Kim, Y, Valente, A.P, Oblatt-Montal, M, Montal, M, Opella, S.J. | | Deposit date: | 2000-04-03 | | Release date: | 2000-04-26 | | Last modified: | 2022-02-16 | | Method: | SOLID-STATE NMR | | Cite: | Structures of the M2 channel-lining segments from nicotinic acetylcholine and NMDA receptors by NMR spectroscopy.
Nat.Struct.Biol., 6, 1999
|
|
2H3O
 
 | | Structure of MERFT, a membrane protein with two trans-membrane helices | | Descriptor: | MerF | | Authors: | Opella, S.J, De Angelis, A.A, Howell, S.C, Nevzorov, A.A. | | Deposit date: | 2006-05-22 | | Release date: | 2006-10-03 | | Last modified: | 2024-05-29 | | Method: | SOLID-STATE NMR | | Cite: | Structure Determination of a Membrane Protein with Two Trans-membrane Helices in Aligned Phospholipid Bicelles by Solid-State NMR Spectroscopy.
J.Am.Chem.Soc., 128, 2006
|
|
1A11
 
 | | NMR STRUCTURE OF MEMBRANE SPANNING SEGMENT 2 OF THE ACETYLCHOLINE RECEPTOR IN DPC MICELLES, 10 STRUCTURES | | Descriptor: | ACETYLCHOLINE RECEPTOR M2 | | Authors: | Gesell, J.J, Sun, W, Montal, M, Opella, S.J. | | Deposit date: | 1997-12-19 | | Release date: | 1998-04-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structures of the M2 channel-lining segments from nicotinic acetylcholine and NMDA receptors by NMR spectroscopy.
Nat.Struct.Biol., 6, 1999
|
|
5WDZ
 
 | |
2KSJ
 
 | | Structure and Dynamics of the Membrane-bound form of Pf1 Coat Protein: Implications for Structural Rearrangement During Virus Assembly | | Descriptor: | Capsid protein G8P | | Authors: | Park, S, Marassi, F, Black, D, Opella, S.J. | | Deposit date: | 2010-01-05 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR, SOLUTION NMR | | Cite: | Structure and dynamics of the membrane-bound form of Pf1 coat protein: implications of structural rearrangement for virus assembly.
Biophys.J., 99, 2010
|
|
1ZN5
 
 | |
1WAZ
 
 | |
1MZT
 
 | |
2JPX
 
 | | A18H Vpu TM structure in lipid bilayers | | Descriptor: | Vpu protein | | Authors: | Park, S, Opella, S.J. | | Deposit date: | 2007-05-25 | | Release date: | 2007-11-13 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Conformational changes induced by a single amino acid substitution in the trans-membrane domain of Vpu: implications for HIV-1 susceptibility to channel blocking drugs
Protein Sci., 16, 2007
|
|
1KVJ
 
 | |
1KVI
 
 | |
1A1U
 
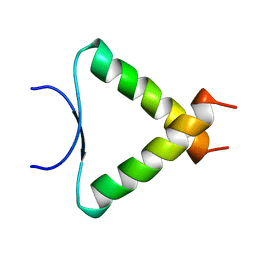 | | SOLUTION STRUCTURE DETERMINATION OF A P53 MUTANT DIMERIZATION DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | P53 | | Authors: | Mccoy, M.A, Stavridi, E.S, Waterman, J.L.F, Wieczorek, A, Opella, S.J, Halezonetis, T.D. | | Deposit date: | 1997-12-16 | | Release date: | 1998-04-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Hydrophobic side-chain size is a determinant of the three-dimensional structure of the p53 oligomerization domain.
EMBO J., 16, 1997
|
|
1AFI
 
 | |
1BKU
 
 | | EFFECTS OF GLYCOSYLATION ON THE STRUCTURE AND DYNAMICS OF EEL CALCITONIN, NMR, 10 STRUCTURES | | Descriptor: | CALCITONIN | | Authors: | Hashimoto, Y, Nishikido, J, Toma, K, Yamamoto, K, Haneda, K, Inazu, T, Valentine, K, Opella, S.J. | | Deposit date: | 1998-07-13 | | Release date: | 1999-01-13 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Effects of glycosylation on the structure and dynamics of eel calcitonin in micelles and lipid bilayers determined by nuclear magnetic resonance spectroscopy.
Biochemistry, 38, 1999
|
|
1AFJ
 
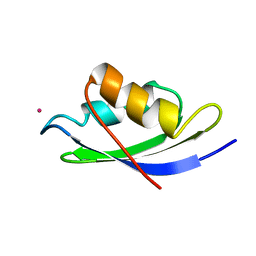 | | STRUCTURE OF THE MERCURY-BOUND FORM OF MERP, THE PERIPLASMIC PROTEIN FROM THE BACTERIAL MERCURY DETOXIFICATION SYSTEM, NMR, 20 STRUCTURES | | Descriptor: | MERCURY (II) ION, MERP | | Authors: | Steele, R.A, Opella, S.J. | | Deposit date: | 1997-03-07 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structures of the reduced and mercury-bound forms of MerP, the periplasmic protein from the bacterial mercury detoxification system.
Biochemistry, 36, 1997
|
|
1BYV
 
 | | GLYCOSYLATED EEL CALCITONIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (CALCITONIN) | | Authors: | Hashimoto, Y, Toma, K, Nishikido, J, Yamamoto, K, Haneda, K, Inazu, T, Valentine, K.G, Opella, S.J. | | Deposit date: | 1998-10-16 | | Release date: | 1998-10-28 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Effects of glycosylation on the structure and dynamics of eel calcitonin in micelles and lipid bilayers determined by nuclear magnetic resonance spectroscopy.
Biochemistry, 38, 1999
|
|
1BZB
 
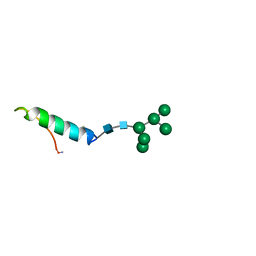 | | GLYCOSYLATED EEL CALCITONIN | | Descriptor: | PROTEIN (CALCITONIN), alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Hashimoto, Y, Toma, K, Nishikido, J, Yamamoto, K, Haneda, K, Inazu, T, Valentine, K, Opella, S.J. | | Deposit date: | 1998-10-27 | | Release date: | 1998-11-11 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Effects of glycosylation on the structure and dynamics of eel calcitonin in micelles and lipid bilayers determined by nuclear magnetic resonance spectroscopy.
Biochemistry, 38, 1999
|
|
2K3M
 
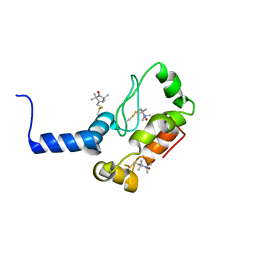 | | Rv1761c | | Descriptor: | Rv1761c, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Page, R.C, Moore, J.D, Lee, S, Opella, S.J, Cross, T.A. | | Deposit date: | 2008-05-14 | | Release date: | 2009-01-06 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Backbone structure of a small helical integral membrane protein: A unique structural characterization.
Protein Sci., 18, 2009
|
|
2GOH
 
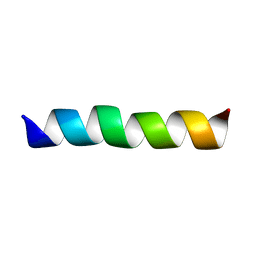 | | Three-dimensional Structure of the Trans-membrane Domain of Vpu from HIV-1 in Aligned Phospholipid Bicelles | | Descriptor: | VPU protein | | Authors: | Park, S.H, De Angelis, A.A, Nevzorov, A.A, Wu, C.H, Opella, S.J. | | Deposit date: | 2006-04-12 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Structure of the Transmembrane Domain of Vpu from HIV-1 in Aligned Phospholipid Bicelles.
Biophys.J., 91, 2006
|
|
2GOF
 
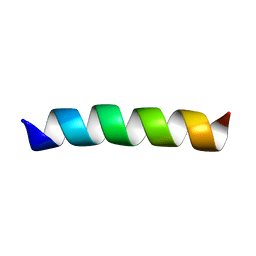 | | Three-dimensional structure of the trans-membrane domain of Vpu from HIV-1 in aligned phospholipid bicelles | | Descriptor: | VPU protein | | Authors: | Park, S.H, De Angelis, A.A, Nevzorov, A.A, Wu, C.H, Opella, S.J. | | Deposit date: | 2006-04-12 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Structure of the Transmembrane Domain of Vpu from HIV-1 in Aligned Phospholipid Bicelles.
Biophys.J., 91, 2006
|
|
2KLV
 
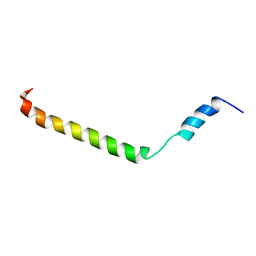 | | Membrane-bound structure of the Pf1 major coat protein in DHPC micelle | | Descriptor: | Capsid protein G8P | | Authors: | Park, S, Son, W, Mukhopadhyay, R, Valafar, H, Opella, S.J. | | Deposit date: | 2009-07-08 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Phage-induced alignment of membrane proteins enables the measurement and structural analysis of residual dipolar couplings with dipolar waves and lambda-maps.
J.Am.Chem.Soc., 131, 2009
|
|
1FDM
 
 | |
1PJD
 
 | | Structure and Topology of a Peptide Segment of the 6th Transmembrane Domain of the Saccharomyces cerevisiae alpha-Factor Receptor in Phospholipid Bilayers | | Descriptor: | Pheromone alpha factor receptor | | Authors: | Valentine, K.G, Liu, S.-F, Marassi, F.M, Veglia, G, Nevzorov, A.A, Opella, S.J, Ding, F.-X, Wang, S.-H, Arshava, B, Becker, J.M, Naider, F. | | Deposit date: | 2003-06-02 | | Release date: | 2003-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLID-STATE NMR | | Cite: | Structure and Topology of a Peptide Segment of the 6th Transmembrane Domain of the Saccharomyces cerevisiae alpha-Factor Receptor in Phospholipid Bilayers
Biopolymers, 59, 2001
|
|
1PJE
 
 | | Structure of the channel-forming trans-membrane domain of Virus protein "u"(Vpu) from HIV-1 | | Descriptor: | VPU protein | | Authors: | Park, S.H, Mrse, A.A, Nevzorov, A.A, Mesleh, M.F, Oblatt-Montal, M, Montal, M, Opella, S.J. | | Deposit date: | 2003-06-02 | | Release date: | 2003-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLID-STATE NMR | | Cite: | Three-dimensional structure of the channel-forming trans-membrane domain of Virus protein "u" (Vpu) from HIV-1
J.Mol.Biol., 333, 2003
|
|
