5FRG
 
 | |
1HK6
 
 | | Ral binding domain from Sec5 | | Descriptor: | EXOCYST COMPLEX COMPONENT SEC5 | | Authors: | Mott, H.R, Nietlispach, D, Hopkins, L.J, Mirey, G, Camonis, J.H, Owen, D. | | Deposit date: | 2003-03-05 | | Release date: | 2003-03-13 | | Last modified: | 2018-03-28 | | Method: | SOLUTION NMR | | Cite: | Structure of the GTPase-binding domain of Sec5 and elucidation of its Ral binding site.
J. Biol. Chem., 278, 2003
|
|
2MBG
 
 | | Rlip76 (gap-gbd) | | Descriptor: | RalA-binding protein 1 | | Authors: | Rajasekar, K.V, Campbell, L.J, Nietlispach, D, Owen, D, Mott, H.R. | | Deposit date: | 2013-07-30 | | Release date: | 2013-12-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Structure of the RLIP76 RhoGAP-Ral Binding Domain Dyad: Fixed Position of the Domains Leads to Dual Engagement of Small G Proteins at the Membrane.
Structure, 21, 2013
|
|
1ZSG
 
 | | beta PIX-SH3 complexed with an atypical peptide from alpha-PAK | | Descriptor: | Rho guanine nucleotide exchange factor 7, Serine/threonine-protein kinase PAK 1 | | Authors: | Mott, H.R, Nietlispach, D, Evetts, K.A, Owen, D. | | Deposit date: | 2005-05-24 | | Release date: | 2005-08-30 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of the SH3 Domain of beta-PIX and Its Interaction with alpha-p21 Activated Kinase (PAK)
Biochemistry, 44, 2005
|
|
2RMK
 
 | | Rac1/PRK1 Complex | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Ras-related C3 botulinum toxin substrate 1, ... | | Authors: | Modha, R, Campbell, L.J, Nietlispach, D, Buhecha, H.R, Owen, D, Mott, H.R. | | Deposit date: | 2007-10-25 | | Release date: | 2007-11-13 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | The Rac1 Polybasic Region Is Required for Interaction with Its Effector PRK1
J.Biol.Chem., 283, 2008
|
|
1URF
 
 | | HR1b domain from PRK1 | | Descriptor: | PROTEIN KINASE C-LIKE 1 | | Authors: | Owen, D, Lowe, P.N, Nietlispach, D, Brosnan, C.E, Chirgadze, D.Y, Parker, P.J, Blundell, T.L, Mott, H.R. | | Deposit date: | 2003-10-29 | | Release date: | 2003-11-06 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Molecular Dissection of the Interaction between the Small G Proteins Rac1 and Rhoa and Protein Kinase C-Related Kinase 1 (Prk1)
J.Biol.Chem., 278, 2003
|
|
1UQV
 
 | | SAM domain from Ste50p | | Descriptor: | STE50 PROTEIN | | Authors: | Grimshaw, S.J, Mott, H.R, Stott, K.M, Nielsen, P.R, Evetts, K.A, Hopkins, L.J, Nietlispach, D, Owen, D. | | Deposit date: | 2003-10-20 | | Release date: | 2003-10-30 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure of the Sterile {Alpha} Motif (Sam) Domain of the Saccharomyces Cerevisiae Mitogen-Activated Protein Kinase Pathway-Modulating Protein Ste50 and Analysis of its Interaction with the Ste11 Sam
J.Biol.Chem., 279, 2004
|
|
5HV8
 
 | | Solution structure of an octanoyl- loaded acyl carrier protein domain from module MLSA2 of the mycolactone polyketide synthase. | | Descriptor: | S-[2-({N-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl] octanethioate, Type I modular polyketide synthase | | Authors: | Vance, S, Tkachenko, O, Thomas, B, Bassuni, M, Hong, H, Nietlispach, D, Broadhurst, R.W. | | Deposit date: | 2016-01-28 | | Release date: | 2016-03-09 | | Last modified: | 2019-10-23 | | Method: | SOLUTION NMR | | Cite: | Sticky swinging arm dynamics: studies of an acyl carrier protein domain from the mycolactone polyketide synthase.
Biochem.J., 473, 2016
|
|
5HVC
 
 | | Solution structure of the apo state of the acyl carrier protein from the MLSA2 subunit of the mycolactone polyketide synthase | | Descriptor: | Type I modular polyketide synthase | | Authors: | Vance, S, Tkachenko, O, Thomas, B, Bassuni, M, Hong, H, Nietlispach, D, Broadhurst, R.W. | | Deposit date: | 2016-01-28 | | Release date: | 2016-03-09 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Sticky swinging arm dynamics: studies of an acyl carrier protein domain from the mycolactone polyketide synthase.
Biochem.J., 473, 2016
|
|
1CF4
 
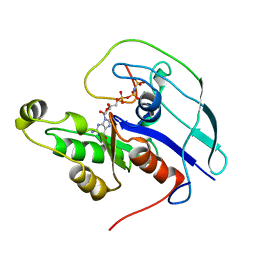 | | CDC42/ACK GTPASE-BINDING DOMAIN COMPLEX | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, PROTEIN (ACTIVATED P21CDC42HS KINASE), ... | | Authors: | Mott, H.R, Owen, D, Nietlispach, D, Lowe, P.N, Lim, L, Laue, E.D. | | Deposit date: | 1999-03-23 | | Release date: | 1999-06-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the small G protein Cdc42 bound to the GTPase-binding domain of ACK.
Nature, 399, 1999
|
|
1E0A
 
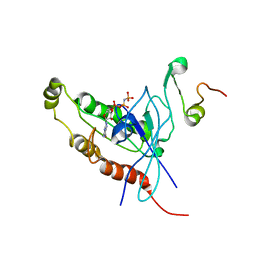 | | Cdc42 complexed with the GTPase binding domain of p21 activated kinase | | Descriptor: | Cell division control protein 42 homolog, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Morreale, A, Venkatesan, M, Mott, H.R, Owen, D, Nietlispach, D, Lowe, P.N, Laue, E.D. | | Deposit date: | 2000-03-16 | | Release date: | 2000-04-18 | | Last modified: | 2019-09-25 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Cdc42 Bound to the Gtpase Binding Domian of Pak
Nat.Struct.Biol., 7, 2000
|
|
1S4Z
 
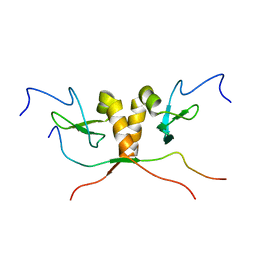 | | HP1 chromo shadow domain in complex with PXVXL motif of CAF-1 | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromobox protein homolog 1 | | Authors: | Thiru, A, Nietlispach, D, Mott, H.R, Okuwaki, M, Lyon, D, Nielsen, P.R, Hirshberg, M, Verreault, A, Murzina, N.V, Laue, E.D. | | Deposit date: | 2004-01-19 | | Release date: | 2004-03-23 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin.
Embo J., 23, 2004
|
|
1H8B
 
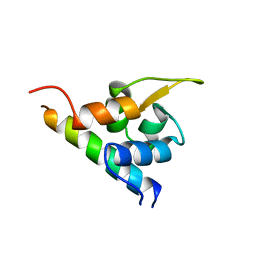 | | EF-hands 3,4 from alpha-actinin / Z-repeat 7 from titin | | Descriptor: | ALPHA-ACTININ 2, SKELETAL MUSCLE ISOFORM, TITIN | | Authors: | Atkinson, R.A, Joseph, C, Kelly, G, Muskett, F.W, Frenkiel, T.A, Nietlispach, D, Pastore, A. | | Deposit date: | 2001-02-01 | | Release date: | 2001-08-30 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Ca2+-Independent Binding of an EF-Hand Domain to a Novel Motif in the Alpha-Actinin-Titin Complex
Nat.Struct.Biol., 8, 2001
|
|
2BUD
 
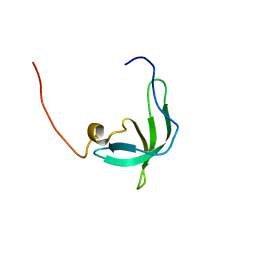 | | The solution structure of the chromo barrel domain from the males- absent on the first (MOF) protein | | Descriptor: | MALES-ABSENT ON THE FIRST PROTEIN | | Authors: | Nielsen, P.R, Nietlispach, D, Buscaino, A, Warner, R.J, Akhtar, A, Murzin, A.G, Murzina, N.V, Laue, E.D. | | Deposit date: | 2005-06-09 | | Release date: | 2005-06-24 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure of the Chromo Barrel Domain from the Mof Acetyltransferase
J.Biol.Chem., 280, 2005
|
|
2BL5
 
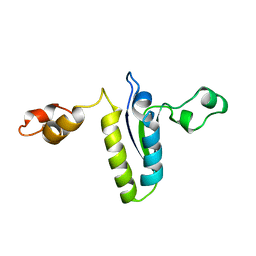 | | Solution structure of the KH-QUA2 region of the Xenopus STAR-GSG Quaking protein. | | Descriptor: | MGC83862 PROTEIN | | Authors: | Maguire, M.L, Guler-Gane, G, Nietlispach, D, Raine, A.R.C, Zorn, A.M, Standart, N, Broadhurst, R.W. | | Deposit date: | 2005-03-01 | | Release date: | 2005-04-14 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Backbone Dynamics of the Kh-Qua2 Region of the Xenopus Star/Gsg Quaking Protein
J.Mol.Biol., 348, 2005
|
|
1XU6
 
 | | Structure of the C-terminal domain from Trypanosoma brucei Variant Surface Glycoprotein MITat1.2 | | Descriptor: | Variant surface glycoprotein MITAT 1.2 | | Authors: | Chattopadhyay, A, Jones, N.G, Nietlispach, D, Nielsen, P.R, Voorheis, H.P, Mott, H.R, Carrington, M. | | Deposit date: | 2004-10-25 | | Release date: | 2004-11-30 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal domain from Trypanosoma brucei variant surface glycoprotein MITat1.2
J.Biol.Chem., 280, 2004
|
|
1DZ1
 
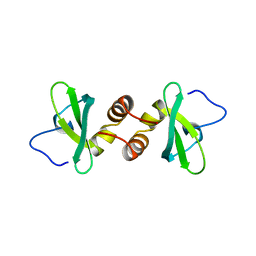 | | Mouse HP1 (M31) C terminal (shadow chromo) domain | | Descriptor: | MODIFIER 1 PROTEIN | | Authors: | Brasher, S.V, Smith, B.O, Fogh, R.H, Nietlispach, D, Thiru, A, Nielsen, P.R, Broadhurst, R.W, Ball, L.J, Murzina, N, Laue, E.D. | | Deposit date: | 2000-02-11 | | Release date: | 2000-04-09 | | Last modified: | 2018-03-07 | | Method: | SOLUTION NMR | | Cite: | The Structure of Mouse Hp1 Suggests a Unique Mode of Single Peptide Recognition by the Shadow Chromo Domain Dimer
Embo J., 19, 2000
|
|
1PZR
 
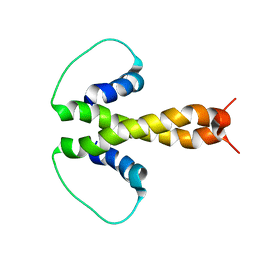 | | Structure of fused docking domains from the erythromycin polyketide synthase (DEBS), a model for the interaction between DEBS2 and DEBS3: the B domain | | Descriptor: | Erythronolide synthase | | Authors: | Broadhurst, R.W, Nietlispach, D, Wheatcroft, M.P, Leadlay, P.F, Weissman, K.J. | | Deposit date: | 2003-07-14 | | Release date: | 2004-02-24 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The structure of docking domains in modular polyketide synthases.
Chem.Biol., 10, 2003
|
|
1PZQ
 
 | | Structure of fused docking domains from the erythromycin polyketide synthase (DEBS), a model for the interaction between DEBS 2 and DEBS 3: The A domain | | Descriptor: | Erythronolide synthase | | Authors: | Broadhurst, R.W, Nietlispach, D, Wheatcroft, M.P, Leadlay, P.F, Weissman, K.J. | | Deposit date: | 2003-07-14 | | Release date: | 2004-02-24 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The structure of docking domains in modular polyketide synthases.
Chem.Biol., 10, 2003
|
|
1GUW
 
 | | STRUCTURE OF THE CHROMODOMAIN FROM MOUSE HP1beta IN COMPLEX WITH THE LYSINE 9-METHYL HISTONE H3 N-TERMINAL PEPTIDE, NMR, 25 STRUCTURES | | Descriptor: | CHROMOBOX PROTEIN HOMOLOG 1, HISTONE H3.1 | | Authors: | Nielsen, P.R, Nietlispach, D, Mott, H.R, Callaghan, J.M, Bannister, A, Kouzarides, T, Murzin, A.G, Murzina, N.V, Laue, E.D. | | Deposit date: | 2002-02-01 | | Release date: | 2002-03-12 | | Last modified: | 2018-01-17 | | Method: | SOLUTION NMR | | Cite: | Structure of the Hp1 Chromodomain Bound to Histone H3 Methylated at Lysine 9
Nature, 416, 2002
|
|
2K9A
 
 | | The Solution Structure of the Arl2 Effector, BART | | Descriptor: | ADP-ribosylation factor-like protein 2-binding protein | | Authors: | Bailey, L.K, Campbell, L.J, Evetts, K.A, Littlefield, K, Rajendra, E, Nietlispach, D, Owen, D, Mott, H.R. | | Deposit date: | 2008-10-06 | | Release date: | 2008-11-11 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The Structure of Binder of Arl2 (BART) Reveals a Novel G Protein Binding Domain: IMPLICATIONS FOR FUNCTION.
J.Biol.Chem., 284, 2009
|
|
2KE5
 
 | | Solution structure and dynamics of the small GTPase Ralb in its active conformation: significance for effector protein binding | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Ral-B | | Authors: | Fenwick, R, Prasannan, S, Campbell, L.J, Nietlispach, D, Evetts, K.A, Camonis, J, Mott, H.R, Owen, D. | | Deposit date: | 2009-01-23 | | Release date: | 2009-02-17 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the small GTPase RalB in its active conformation: significance for effector protein binding
Biochemistry, 48, 2009
|
|
2KWH
 
 | | Ral binding domain of RLIP76 (RalBP1) | | Descriptor: | RalA-binding protein 1 | | Authors: | Fenwick, R.B, Campbell, L.J, Rajasekar, K, Prasannan, S, Nietlispach, D, Camonis, J, Owen, D, Mott, H.R. | | Deposit date: | 2010-04-12 | | Release date: | 2010-09-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The RalB-RLIP76 complex reveals a novel mode of ral-effector interaction
Structure, 18, 2010
|
|
2KWI
 
 | | RalB-RLIP76 (RalBP1) complex | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, RalA-binding protein 1, ... | | Authors: | Fenwick, R.B, Campbell, L.J, Rajasekar, K, Prasannan, S, Nietlispach, D, Camonis, J, Owen, D, Mott, H.R. | | Deposit date: | 2010-04-12 | | Release date: | 2010-09-01 | | Last modified: | 2021-10-06 | | Method: | SOLUTION NMR | | Cite: | The RalB-RLIP76 complex reveals a novel mode of ral-effector interaction
Structure, 18, 2010
|
|
2JWH
 
 | | Structure of a Glycosylphosphatidylinositol-anchored Domain from a Trypanosome Variant Surface Glycoprotein | | Descriptor: | Variant surface glycoprotein ILTAT 1.24 | | Authors: | Jones, N.G, Nietlispach, D, Sharma, R, Burke, D.F, Eyres, I, Mues, M, Mott, H.R, Carrington, M. | | Deposit date: | 2007-10-12 | | Release date: | 2007-11-13 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure of a Glycosylphosphatidylinositol-anchored Domain from a Trypanosome Variant Surface Glycoprotein
J.Biol.Chem., 283, 2008
|
|
