1AHZ
 
 | |
1AHV
 
 | |
1AHU
 
 | |
3LAD
 
 | |
1VAO
 
 | | STRUCTURE OF THE OCTAMERIC FLAVOENZYME VANILLYL-ALCOHOL OXIDASE | | Descriptor: | ACETATE ION, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mattevi, A. | | Deposit date: | 1997-04-10 | | Release date: | 1997-10-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures and inhibitor binding in the octameric flavoenzyme vanillyl-alcohol oxidase: the shape of the active-site cavity controls substrate specificity.
Structure, 5, 1997
|
|
1EAE
 
 | |
1EAA
 
 | |
1EAD
 
 | |
1EAB
 
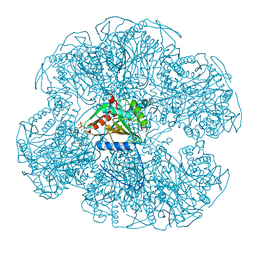 | |
1EAF
 
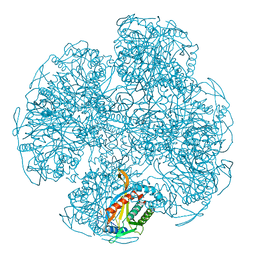 | |
1EAC
 
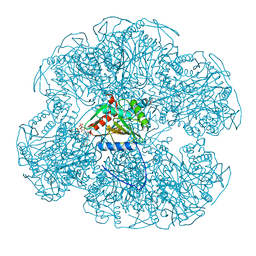 | |
1LPF
 
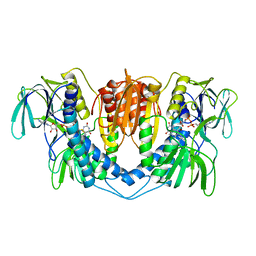 | |
1PKY
 
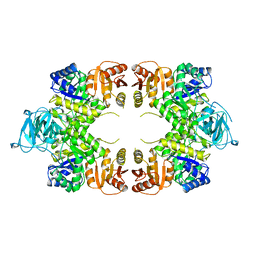 | | PYRUVATE KINASE FROM E. COLI IN THE T-STATE | | Descriptor: | PYRUVATE KINASE | | Authors: | Mattevi, A. | | Deposit date: | 1995-04-27 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Escherichia coli pyruvate kinase type I: molecular basis of the allosteric transition.
Structure, 3, 1995
|
|
1CHU
 
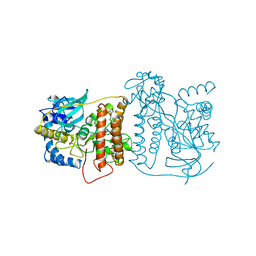 | |
1LVL
 
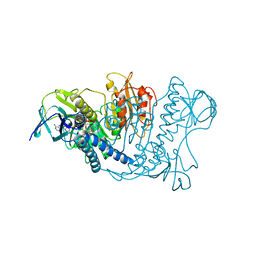 | |
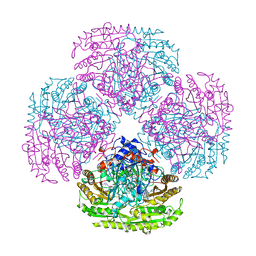
2VAO
 
 | |
6TUY
 
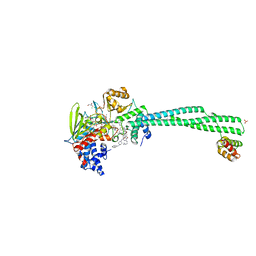 | | Human LSD1/CoREST bound to the quinazoline inhibitor MC4106 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mattevi, A, Marrocco, B. | | Deposit date: | 2020-01-08 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel non-covalent LSD1 inhibitors endowed with anticancer effects in leukemia and solid tumor cellular models.
Eur.J.Med.Chem., 237, 2022
|
|
6GOU
 
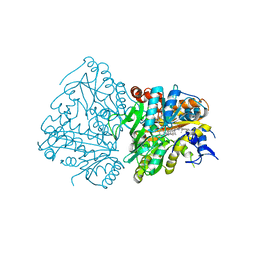 | | Development of Alkyl Glycerone Phosphate Synthase Inhibitors: Complex with Inhibitor 2I | | Descriptor: | (3~{S})-3-[2,6-bis(fluoranyl)phenyl]-~{N}-[(2-oxidanylidene-1,3-dihydrobenzimidazol-5-yl)methyl]butanamide, Alkyldihydroxyacetonephosphate synthase, peroxisomal, ... | | Authors: | Mattevi, A, Piano, V. | | Deposit date: | 2018-06-04 | | Release date: | 2019-01-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Development of alkyl glycerone phosphate synthase inhibitors: Structure-activity relationship and effects on ether lipids and epithelial-mesenchymal transition in cancer cells.
Eur J Med Chem, 163, 2018
|
|
1QLT
 
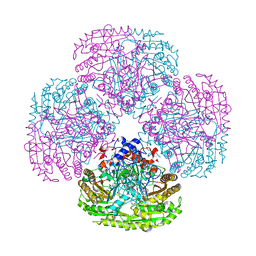 | |
5LGT
 
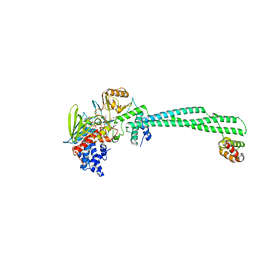 | | Thieno[3,2-b]pyrrole-5-carboxamides as Novel Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1: Compound 15 | | Descriptor: | 4-methyl-~{N}-[2-[[4-(1-methylpiperidin-4-yl)oxyphenoxy]methyl]phenyl]thieno[3,2-b]pyrrole-5-carboxamide, FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, ... | | Authors: | Mattevi, A, Ciossani, G. | | Deposit date: | 2016-07-08 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Thieno[3,2-b]pyrrole-5-carboxamides as New Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1. Part 2: Structure-Based Drug Design and Structure-Activity Relationship.
J. Med. Chem., 60, 2017
|
|
5LGU
 
 | | Thieno[3,2-b]pyrrole-5-carboxamides as Novel Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1: Compound 34 | | Descriptor: | 4-methyl-~{N}-[2-[[4-[[(3~{R})-pyrrolidin-3-yl]methoxy]phenoxy]methyl]phenyl]thieno[3,2-b]pyrrole-5-carboxamide, FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, ... | | Authors: | Mattevi, A, Ciossani, G. | | Deposit date: | 2016-07-08 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Thieno[3,2-b]pyrrole-5-carboxamides as New Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1. Part 2: Structure-Based Drug Design and Structure-Activity Relationship.
J. Med. Chem., 60, 2017
|
|
5LGN
 
 | | Thieno[3,2-b]pyrrole-5-carboxamides as Novel Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1: Compound 19 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, REST corepressor 1, ... | | Authors: | Mattevi, A, Ciossani, G. | | Deposit date: | 2016-07-07 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Thieno[3,2-b]pyrrole-5-carboxamides as New Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1. Part 1: High-Throughput Screening and Preliminary Exploration.
J. Med. Chem., 60, 2017
|
|
1QLU
 
 | |
6GQI
 
 | |
1KIF
 
 | | D-AMINO ACID OXIDASE FROM PIG KIDNEY | | Descriptor: | BENZOIC ACID, D-AMINO ACID OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Todone, F, Mattevi, A. | | Deposit date: | 1996-01-19 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of D-amino acid oxidase: a case of active site mirror-image convergent evolution with flavocytochrome b2.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
