3E5F
 
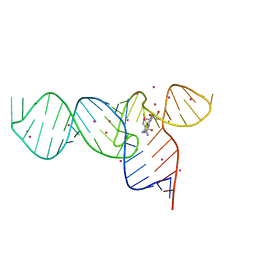 | | Crystal Structures of the SMK box (SAM-III) Riboswitch with Se-SAM | | Descriptor: | SMK box (SAM-III) Riboswitch for RNA, STRONTIUM ION, [(3S)-3-amino-4-hydroxy-4-oxo-butyl]-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methyl]-methyl-selanium | | Authors: | Lu, C. | | Deposit date: | 2008-08-13 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of the SAM-III/S(MK) riboswitch reveal the SAM-dependent translation inhibition mechanism.
Nat.Struct.Mol.Biol., 15, 2008
|
|
3E5C
 
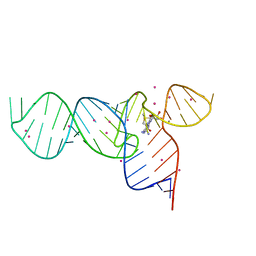 | | Crystal Structure of the SMK box (SAM-III) Riboswitch with SAM | | Descriptor: | S-ADENOSYLMETHIONINE, SMK box (SAM-III) Riboswitch, STRONTIUM ION | | Authors: | Lu, C. | | Deposit date: | 2008-08-13 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of the SAM-III/S(MK) riboswitch reveal the SAM-dependent translation inhibition mechanism.
Nat.Struct.Mol.Biol., 15, 2008
|
|
3E5E
 
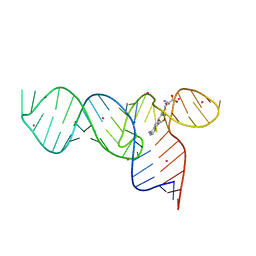 | | Crystal Structures of the SMK box (SAM-III) Riboswitch with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SMK box (SAM-III) Riboswitch for RNA, STRONTIUM ION | | Authors: | Lu, C. | | Deposit date: | 2008-08-13 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of the SAM-III/S(MK) riboswitch reveal the SAM-dependent translation inhibition mechanism.
Nat.Struct.Mol.Biol., 15, 2008
|
|
7VIR
 
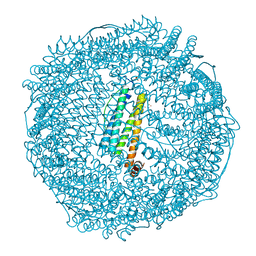 | | Crystal structure of Au(100EQ)-apo-R168H/L169C-rHLFr | | Descriptor: | CADMIUM ION, Ferritin light chain, GOLD ION, ... | | Authors: | Lu, C, Peng, X, Maity, B, Ito, N, Abe, S, Ueno, T, Lu, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of a gold clustering site in an engineered apo-ferritin cage.
Commun Chem, 5, 2022
|
|
7VIO
 
 | | Crystal structure of recombinant horse spleen apo-R168H/L169C-Fr | | Descriptor: | CADMIUM ION, Ferritin light chain, GLYCEROL, ... | | Authors: | Lu, C, Peng, X, Maity, B, Ito, N, Abe, S, Wu, J, Ueno, T, Lu, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of a gold clustering site in an engineered apo-ferritin cage.
Commun Chem, 5, 2022
|
|
7VIU
 
 | | Crystal structure of Au(200EQ)-apo-R168C/L169C-rHLFr | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ferritin light chain, ... | | Authors: | Lu, C, Peng, X, Maity, B, Ito, N, Abe, S, Ueno, T, Lu, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of a gold clustering site in an engineered apo-ferritin cage.
Commun Chem, 5, 2022
|
|
7VIT
 
 | | Crystal structure of Au(400EQ)-apo-R168H/L169C-rHLFr | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ferritin light chain, ... | | Authors: | Lu, C, Peng, X, Maity, B, Ito, N, Abe, S, Ueno, T, Lu, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of a gold clustering site in an engineered apo-ferritin cage.
Commun Chem, 5, 2022
|
|
7VIQ
 
 | | Crystal structure of Au(50EQ)-apo-R168H/L169C-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Lu, C, Peng, X, Maity, B, Ito, N, Abe, S, Ueno, T, Lu, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-06-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of a gold clustering site in an engineered apo-ferritin cage.
Commun Chem, 5, 2022
|
|
7VIP
 
 | | Crystal structure of Au(10EQ)-apo-R168H/L169C-rHLFr | | Descriptor: | CADMIUM ION, Ferritin light chain, GLYCEROL, ... | | Authors: | Lu, C, Peng, X, Maity, B, Ito, N, Abe, S, Ueno, T, Lu, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of a gold clustering site in an engineered apo-ferritin cage.
Commun Chem, 5, 2022
|
|
7VIS
 
 | | Crystal structure of Au(200EQ)-apo-R168H/L169C-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Lu, C, Peng, X, Maity, B, Ito, N, Abe, S, Ueno, T, Lu, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design of a gold clustering site in an engineered apo-ferritin cage.
Commun Chem, 5, 2022
|
|
3L7Z
 
 | | Crystal structure of the S. solfataricus archaeal exosome | | Descriptor: | Probable exosome complex RNA-binding protein 1, Probable exosome complex exonuclease 1, Probable exosome complex exonuclease 2, ... | | Authors: | Lu, C, Ding, F, Ke, A. | | Deposit date: | 2009-12-29 | | Release date: | 2010-06-23 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structure of the S. solfataricus archaeal exosome reveals conformational flexibility in the RNA-binding ring.
Plos One, 5, 2010
|
|
3NJP
 
 | | The Extracellular and Transmembrane Domain Interfaces in Epidermal Growth Factor Receptor Signaling | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor, Epidermal growth factor receptor, ... | | Authors: | Lu, C, Mi, L.-Z, Grey, M.J, Zhu, J, Graef, E, Yokoyama, S, Springer, T.A. | | Deposit date: | 2010-06-17 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.304 Å) | | Cite: | Structural evidence for loose linkage between ligand binding and kinase activation in the epidermal growth factor receptor.
Mol.Cell.Biol., 30, 2010
|
|
4KQY
 
 | | Bacillus subtilis yitJ S box/SAM-I riboswitch | | Descriptor: | MAGNESIUM ION, S-ADENOSYLMETHIONINE, YitJ S box/SAM-I riboswitch | | Authors: | Lu, C. | | Deposit date: | 2013-05-15 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | SAM recognition and conformational switching mechanism in the Bacillus subtilis yitJ S box/SAM-I riboswitch
J.Mol.Biol., 404, 2010
|
|
4PHT
 
 | | ATPase GspE in complex with the cytoplasmic domain of GspL from the Vibrio vulnificus type II Secretion system | | Descriptor: | General secretory pathway protein E, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Lu, C, Korotkov, K, Hol, W. | | Deposit date: | 2014-05-06 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Crystal structure of the full-length ATPase GspE from the Vibrio vulnificus type II secretion system in complex with the cytoplasmic domain of GspL.
J.Struct.Biol., 187, 2014
|
|
4XCH
 
 | |
3VDJ
 
 | | Crystal structure of circumsporozoite protein aTSR domain, R32 native form | | Descriptor: | Circumsporozoite (CS) protein | | Authors: | Doud, M.B, Koksal, A.C, Mi, L.Z, Song, G, Lu, C, Springer, T.A. | | Deposit date: | 2012-01-05 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Unexpected fold in the circumsporozoite protein target of malaria vaccines.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VDK
 
 | | Crystal structure of circumsporozoite protein aTSR domain, R32 platinum-bound form | | Descriptor: | Circumsporozoite (CS) protein, PLATINUM (II) ION | | Authors: | Doud, M.B, Koksal, A.C, Mi, L.Z, Song, G, Lu, C, Springer, T.A. | | Deposit date: | 2012-01-05 | | Release date: | 2012-05-09 | | Last modified: | 2012-05-30 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Unexpected fold in the circumsporozoite protein target of malaria vaccines.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VDL
 
 | | Crystal structure of circumsporozoite protein aTSR domain, P43212 form | | Descriptor: | Circumsporozoite (CS) protein | | Authors: | Doud, M.B, Koksal, A.C, Mi, L.Z, Song, G, Lu, C, Springer, T.A. | | Deposit date: | 2012-01-05 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Unexpected fold in the circumsporozoite protein target of malaria vaccines.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3R4F
 
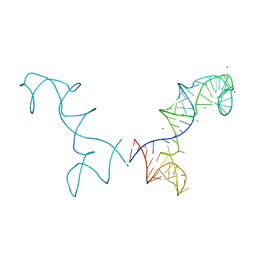 | | Prohead RNA | | Descriptor: | MAGNESIUM ION, pRNA | | Authors: | Ding, F, Lu, C, Zhano, W, Rajashankar, K.R, Anderson, D.L, Jardine, P.J, Grimes, S, Ke, A. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and assembly of the essential RNA ring component of a viral DNA packaging motor.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6E35
 
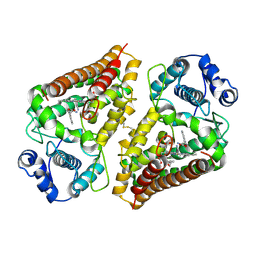 | | Crystal structure of human indoleamime 2,3-dioxygenase (IDO1) in complex with L-Trp and cyanide, Northeast Structural Genomics Target HR6160 | | Descriptor: | 2-(1H-indol-3-yl)ethanol, CYANIDE ION, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Forouhar, F, Lewis-Ballester, A, Lew, S, Karkashon, S, Seetharaman, J, Lu, C, Hussain, M, Yeh, S.-R, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2018-07-13 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.411 Å) | | Cite: | Crystal structure of human indoleamime 2,3-dioxygenase (IDO1) in complex with L-Trp and cyanide, Northeast Structural Genomics Target HR6160
To Be Published
|
|
6JEE
 
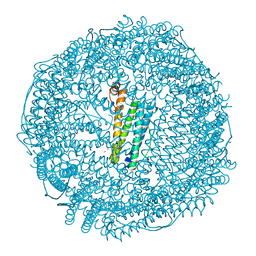 | | Crystal structure of apo-L161C/L165C-Fr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Abe, S, Ito, N, Maity, B, Lu, C, Lu, D, Ueno, T. | | Deposit date: | 2019-02-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Coordination design of cadmium ions at the 4-fold axis channel of the apo-ferritin cage.
Dalton Trans, 48, 2019
|
|
6JEF
 
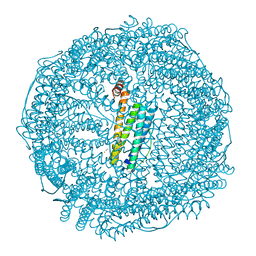 | | Crystal structure of apo-R168C/L169C-Fr | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ferritin light chain, ... | | Authors: | Abe, S, Ito, N, Maity, B, Lu, C, Lu, D, Ueno, T. | | Deposit date: | 2019-02-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Coordination design of cadmium ions at the 4-fold axis channel of the apo-ferritin cage.
Dalton Trans, 48, 2019
|
|
4HQO
 
 | | Crystal structure of Plasmodium vivax TRAP protein | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Song, G, Koksal, A.C, Lu, C, Springer, T.A. | | Deposit date: | 2012-10-25 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Shape change in the receptor for gliding motility in Plasmodium sporozoites.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4HQL
 
 | | Crystal structure of magnesium-loaded Plasmodium vivax TRAP protein | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Sporozoite surface protein 2, ... | | Authors: | Song, G, Koksal, A.C, Lu, C, Springer, T.A. | | Deposit date: | 2012-10-25 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.241 Å) | | Cite: | Shape change in the receptor for gliding motility in Plasmodium sporozoites.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4HQK
 
 | | Crystal structure of Plasmodium falciparum TRAP, P4212 form | | Descriptor: | SULFATE ION, Thrombospondin-related anonymous protein, TRAP | | Authors: | Song, G, Koksal, A.C, Lu, C, Springer, T.A. | | Deposit date: | 2012-10-25 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Shape change in the receptor for gliding motility in Plasmodium sporozoites.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
