2IGZ
 
 | |
1B45
 
 | | ALPHA-CNIA CONOTOXIN FROM CONUS CONSORS, NMR, 43 STRUCTURES | | Descriptor: | ALPHA-CNIA | | Authors: | Favreau, P, Krimm, I, Le Gall, F, Bobenrieth, M.J, Lamthanh, H, Bouet, F, Servent, D, Molgo, J, Menez, A, Letourneux, Y, Lancelin, J.M. | | Deposit date: | 1999-01-05 | | Release date: | 1999-07-09 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Biochemical characterization and nuclear magnetic resonance structure of novel alpha-conotoxins isolated from the venom of Conus consors.
Biochemistry, 38, 1999
|
|
2NPV
 
 | |
1FH3
 
 | |
1TP9
 
 | | PRX D (type II) from Populus tremula | | Descriptor: | SULFATE ION, peroxiredoxin | | Authors: | Echalier, A, Trivelli, X, Corbier, C, Rouhier, N, Walker, O, Tsan, P, Jacquot, J.P, Krimm, I, Lancelin, J.M. | | Deposit date: | 2004-06-16 | | Release date: | 2005-04-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure and solution NMR dynamics of a D (type II) peroxiredoxin glutaredoxin and thioredoxin dependent: a new insight into the peroxiredoxin oligomerism
Biochemistry, 44, 2005
|
|
1G2G
 
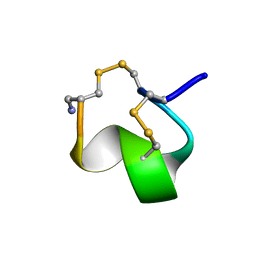 | | MINIMAL CONFORMATION OF THE ALPHA-CONOTOXIN IMI FOR THE ALPHA7 NEURONAL NICOTINIC ACETYLCHOLINE RECEPTOR RECOGNITION | | Descriptor: | ALPHA-CONOTOXIN IMI | | Authors: | Lamthanh, H, Jegou-Matheron, C, Servent, D, Menez, A, Lancelin, J.M. | | Deposit date: | 2000-10-19 | | Release date: | 2000-11-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Minimal conformation of the alpha-conotoxin ImI for the alpha7 neuronal nicotinic acetylcholine receptor recognition: correlated CD, NMR and binding studies.
FEBS Lett., 454, 1999
|
|
1G1Z
 
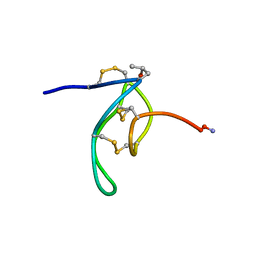 | | NMR Solution Structures of delta-Conotoxin EVIA from Conus ermineus that Selectively Acts on Vertebrate Neuronal Na+ Channels, LEU12-PRO13 Cis isomer | | Descriptor: | CONOTOXIN EVIA | | Authors: | Volpon, L, Lamthanh, H, Le Gall, F, Menez, A, Lancelin, J.M. | | Deposit date: | 2000-10-16 | | Release date: | 2000-11-01 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structures of delta-Conotoxin EVIA from Conus ermineus That Selectively Acts on Vertebrate Neuronal Na+ Channels.
J.Biol.Chem., 279, 2004
|
|
1G1P
 
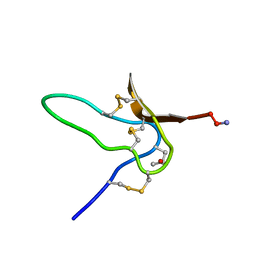 | | NMR Solution Structures of delta-Conotoxin EVIA from Conus ermineus that Selectively Acts on Vertebrate Neuronal Na+ Channels | | Descriptor: | CONOTOXIN EVIA | | Authors: | Volpon, L, Lamthanh, H, Barbier, J, Gilles, N, Molgo, J, Menez, A, Lancelin, J.M. | | Deposit date: | 2000-10-13 | | Release date: | 2000-11-01 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structures of delta-Conotoxin EVIA from Conus ermineus That Selectively Acts on Vertebrate Neuronal Na+ Channels.
J.Biol.Chem., 279, 2004
|
|
1DBY
 
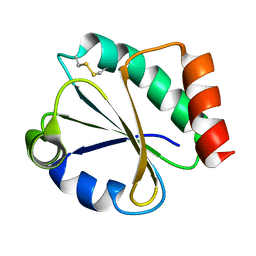 | | NMR STRUCTURES OF CHLOROPLAST THIOREDOXIN M CH2 FROM THE GREEN ALGA CHLAMYDOMONAS REINHARDTII | | Descriptor: | CHLOROPLAST THIOREDOXIN M CH2 | | Authors: | Lancelin, J.-M, Guilhaudis, L, Krimm, I, Blackledge, M.J, Marion, D. | | Deposit date: | 1999-11-03 | | Release date: | 1999-11-08 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR structures of thioredoxin m from the green alga Chlamydomonas reinhardtii.
Proteins, 41, 2000
|
|
1DEN
 
 | |
1DEM
 
 | |
1FCT
 
 | |
1TOF
 
 | | THIOREDOXIN H (OXIDIZED FORM), NMR, 23 STRUCTURES | | Descriptor: | THIOREDOXIN H | | Authors: | Mittard, V, Blackledge, M.J, Stein, M, Jacquot, J.-P, Marion, D, Lancelin, J.-M. | | Deposit date: | 1996-05-30 | | Release date: | 1996-12-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of an oxidised thioredoxin h from the eukaryotic green alga Chlamydomonas reinhardtii.
Eur.J.Biochem., 243, 1997
|
|
2IH0
 
 | |
1BMR
 
 | | ALPHA-LIKE TOXIN LQH III FROM SCORPION LEIURUS QUINQUESTRIATUS HEBRAEUS, NMR, 25 STRUCTURES | | Descriptor: | LQH III ALPHA-LIKE TOXIN | | Authors: | Krimm, I, Gilles, N, Sautiere, P, Stankiewicz, M, Pelhate, M, Gordon, D, Lancelin, J.-M. | | Deposit date: | 1998-07-24 | | Release date: | 1999-02-16 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR structures and activity of a novel alpha-like toxin from the scorpion Leiurus quinquestriatus hebraeus.
J.Mol.Biol., 285, 1999
|
|
1TCJ
 
 | |
1TCG
 
 | |
1TCK
 
 | |
1TCH
 
 | |
2L0T
 
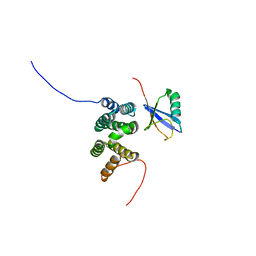 | | Solution structure of the complex of ubiquitin and the VHS domain of Stam2 | | Descriptor: | Signal transducing adapter molecule 2, Ubiquitin | | Authors: | Lange, A, Hoeller, D, Wienk, H, Marcillat, O, Lancelin, J, Walker, O. | | Deposit date: | 2010-07-15 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR reveals a different mode of binding of the Stam2 VHS domain to ubiquitin and diubiquitin.
Biochemistry, 50, 2011
|
|
1GIB
 
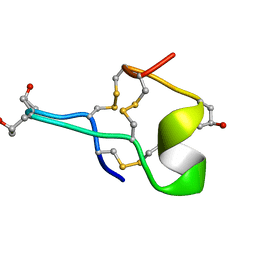 | | MU-CONOTOXIN GIIIB, NMR | | Descriptor: | MU-CONOTOXIN GIIIB | | Authors: | Hill, J.M, Alewood, P.F, Craik, D.J. | | Deposit date: | 1996-04-17 | | Release date: | 1996-11-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of mu-conotoxin GIIIB, a specific blocker of skeletal muscle sodium channels.
Biochemistry, 35, 1996
|
|
2MOA
 
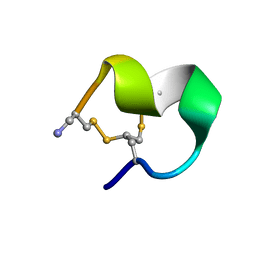 | |
1EP7
 
 | | CRYSTAL STRUCTURE OF WT THIOREDOXIN H FROM CHLAMYDOMONAS REINHARDTII | | Descriptor: | THIOREDOXIN CH1, H-TYPE | | Authors: | Menchise, V, Corbier, C, Didierjean, C, Saviano, M, Benedetti, E, Jacquot, J.P, Aubry, A. | | Deposit date: | 2000-03-28 | | Release date: | 2001-12-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the wild-type and D30A mutant thioredoxin h of Chlamydomonas reinhardtii and implications for the catalytic mechanism.
Biochem.J., 359, 2001
|
|
1EP8
 
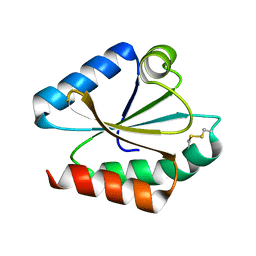 | | CRYSTAL STRUCTURE OF A MUTATED THIOREDOXIN, D30A, FROM CHLAMYDOMONAS REINHARDTII | | Descriptor: | THIOREDOXIN CH1, H-TYPE | | Authors: | Menchise, V, Corbier, C, Didierjean, C, Saviano, M, Benedetti, E, Jacquot, J.P, Aubry, A. | | Deposit date: | 2000-03-28 | | Release date: | 2001-12-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the wild-type and D30A mutant thioredoxin h of Chlamydomonas reinhardtii and implications for the catalytic mechanism.
Biochem.J., 359, 2001
|
|
