1GRB
 
 | |
1GRG
 
 | |
1GRF
 
 | |
1GRE
 
 | |
1GRA
 
 | |
1GRH
 
 | | INHIBITION OF HUMAN GLUTATHIONE REDUCTASE BY THE NITROSOUREA DRUGS 1,3-BIS(2-CHLOROETHYL)-1-NITROSOUREA AND 1-(2-CHLOROETHYL)-3-(2-HYDROXYETHYL)-1-NITROSOUREA | | Descriptor: | ETHANOL, FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE, ... | | Authors: | Karplus, P.A, Schulz, G.E. | | Deposit date: | 1992-12-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibition of human glutathione reductase by the nitrosourea drugs 1,3-bis(2-chloroethyl)-1-nitrosourea and 1-(2-chloroethyl)-3-(2-hydroxyethyl)-1-nitrosourea. A crystallographic analysis.
Eur.J.Biochem., 171, 1988
|
|
3GRS
 
 | |
3EMP
 
 | |
3LVB
 
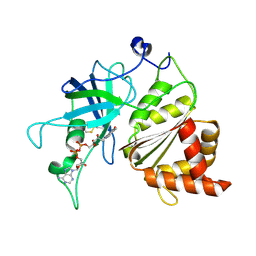 | | Crystal structure of the Ferredoxin:NADP+ reductase from maize root at 1.7 angstroms - Test Set Withheld | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin-NADP reductase | | Authors: | Faber, H.R, Karplus, P.A, Aliverti, A, Ferioli, C, Spinola, M. | | Deposit date: | 2010-02-19 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and crystallographic characterization of ferredoxin-NADP(+) reductase from nonphotosynthetic tissues
Biochemistry, 40, 2001
|
|
2RGO
 
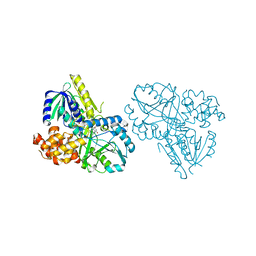 | | Structure of Alpha-Glycerophosphate Oxidase from Streptococcus sp.: A Template for the Mitochondrial Alpha-Glycerophosphate Dehydrogenase | | Descriptor: | Alpha-Glycerophosphate Oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Colussi, T, Boles, W, Mallett, T.C, Karplus, P.A, Claiborne, A. | | Deposit date: | 2007-10-04 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of alpha-glycerophosphate oxidase from Streptococcus sp.: a template for the mitochondrial alpha-glycerophosphate dehydrogenase.
Biochemistry, 47, 2008
|
|
1EJX
 
 | |
3DK8
 
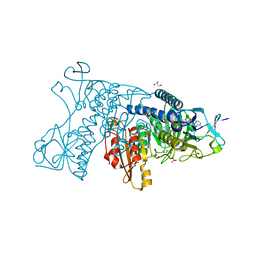 | | Catalytic cycle of human glutathione reductase near 1 A resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE, GLYCEROL, ... | | Authors: | Berkholz, D.S, Faber, H.R, Savvides, S.N, Karplus, P.A. | | Deposit date: | 2008-06-24 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Catalytic cycle of human glutathione reductase near 1 A resolution.
J.Mol.Biol., 382, 2008
|
|
2GMF
 
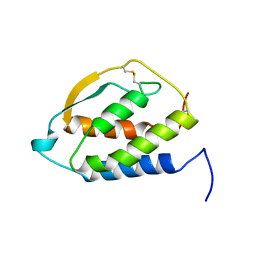 | | HUMAN GRANULOCYTE MACROPHAGE COLONY STIMULATING FACTOR | | Descriptor: | GRANULOCYTE-MACROPHAGE COLONY-STIMULATING FACTOR | | Authors: | Rozwarski, D, Diederichs, K, Hecht, R, Boone, T, Karplus, P.A. | | Deposit date: | 1996-04-24 | | Release date: | 1996-11-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Refined crystal structure and mutagenesis of human granulocyte-macrophage colony-stimulating factor.
Proteins, 26, 1996
|
|
3LO8
 
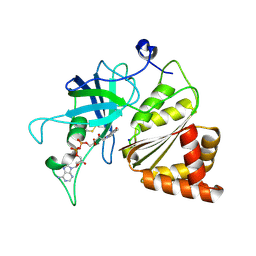 | |
3MNG
 
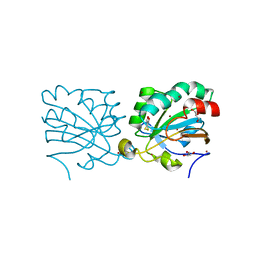 | | wild type human PrxV with DTT bound as a competitive inhibitor | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, BROMIDE ION, GLYCEROL, ... | | Authors: | Hall, A, Karplus, P.A. | | Deposit date: | 2010-04-21 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Evidence that Peroxiredoxin Catalytic Power Is Based on Transition-State Stabilization.
J.Mol.Biol., 402, 2010
|
|
4P53
 
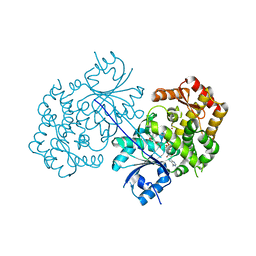 | | ValA (2-epi-5-epi-valiolone synthase) from Streptomyces hygroscopicus subsp. jinggangensis 5008 with NAD+ and Zn2+ bound | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Cyclase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kean, K.M, Codding, S.J, Karplus, P.A. | | Deposit date: | 2014-03-13 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a sedoheptulose 7-phosphate cyclase: ValA from Streptomyces hygroscopicus.
Biochemistry, 53, 2014
|
|
4PBT
 
 | | Crystal structure of the M. jannaschii G2 tRNA synthetase variant bound to 4-trans-cyclooctene-amidopheylalanine (Tco-amF) | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 4-{[(1R,4E)-cyclooct-4-en-1-ylcarbonyl]amino}-L-phenylalanine, GLYCEROL, ... | | Authors: | Cooley, R.B, Karplus, P.A, Mehl, R.A. | | Deposit date: | 2014-04-13 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Gleaning Unexpected Fruits from Hard-Won Synthetases: Probing Principles of Permissivity in Non-canonical Amino Acid-tRNA Synthetases.
Chembiochem, 15, 2014
|
|
4PJY
 
 | | Azide bound Cysteine Dioxygenase at pH 6.2 | | Descriptor: | AZIDE ION, Cysteine dioxygenase type 1, FE (III) ION | | Authors: | Driggers, C.M, Karplus, P.A. | | Deposit date: | 2014-05-12 | | Release date: | 2016-02-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Structure-Based Insights into the Role of the Cys-Tyr Crosslink and Inhibitor Recognition by Mammalian Cysteine Dioxygenase.
J. Mol. Biol., 428, 2016
|
|
4PBS
 
 | |
4PTZ
 
 | | Crystal structure of the Escherichia coli alkanesulfonate FMN reductase SsuE in FMN-bound form | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN reductase SsuE, GLYCEROL, ... | | Authors: | Driggers, C.M, Ellis, H.R, Karplus, P.A. | | Deposit date: | 2014-03-11 | | Release date: | 2014-06-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9007 Å) | | Cite: | Crystal Structure of Escherichia coli SsuE: Defining a General Catalytic Cycle for FMN Reductases of the Flavodoxin-like Superfamily.
Biochemistry, 53, 2014
|
|
6DVI
 
 | |
6B00
 
 | |
6DVH
 
 | |
5I0T
 
 | | Thiosulfate bound Cysteine Dioxygenase at pH 6.8 | | Descriptor: | Cysteine dioxygenase type 1, FE (III) ION, THIOSULFATE | | Authors: | Kean, K.M, Driggers, C.M, Karplus, P.A. | | Deposit date: | 2016-02-04 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structure-Based Insights into the Role of the Cys-Tyr Crosslink and Inhibitor Recognition by Mammalian Cysteine Dioxygenase.
J. Mol. Biol., 428, 2016
|
|
5I0S
 
 | | Thiosulfate bound Cysteine Dioxygenase at pH 6.2 | | Descriptor: | Cysteine dioxygenase type 1, FE (III) ION, THIOSULFATE | | Authors: | Kean, K.M, Driggers, C.M, Karplus, P.A. | | Deposit date: | 2016-02-04 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-Based Insights into the Role of the Cys-Tyr Crosslink and Inhibitor Recognition by Mammalian Cysteine Dioxygenase.
J. Mol. Biol., 428, 2016
|
|
