1GRB
 
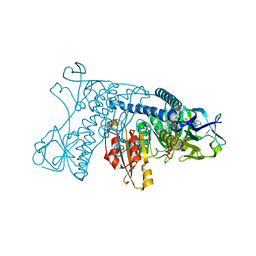 | |
1GRG
 
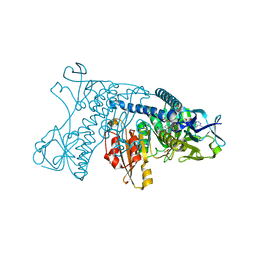 | |
1GRF
 
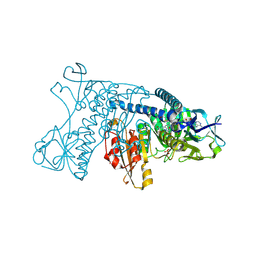 | |
1GRA
 
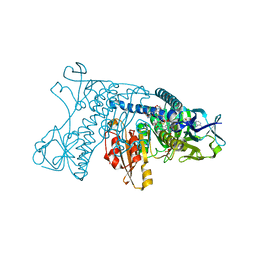 | |
1GRE
 
 | |
1GRH
 
 | | INHIBITION OF HUMAN GLUTATHIONE REDUCTASE BY THE NITROSOUREA DRUGS 1,3-BIS(2-CHLOROETHYL)-1-NITROSOUREA AND 1-(2-CHLOROETHYL)-3-(2-HYDROXYETHYL)-1-NITROSOUREA | | Descriptor: | ETHANOL, FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE, ... | | Authors: | Karplus, P.A, Schulz, G.E. | | Deposit date: | 1992-12-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibition of human glutathione reductase by the nitrosourea drugs 1,3-bis(2-chloroethyl)-1-nitrosourea and 1-(2-chloroethyl)-3-(2-hydroxyethyl)-1-nitrosourea. A crystallographic analysis.
Eur.J.Biochem., 171, 1988
|
|
3GRS
 
 | |
3EMP
 
 | |
1KRA
 
 | |
1KRB
 
 | |
1KRC
 
 | |
3SID
 
 | | Crystal structure of oxidized Symerythrin from Cyanophora paradoxa, azide adduct at 50% occupancy | | Descriptor: | AZIDE ION, FE (III) ION, Symerythrin | | Authors: | Cooley, R.B, Arp, D.J, Karplus, P.A. | | Deposit date: | 2011-06-17 | | Release date: | 2011-08-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Symerythrin structures at atomic resolution and the origins of rubrerythrins and the ferritin-like superfamily.
J.Mol.Biol., 413, 2011
|
|
2KAU
 
 | | THE CRYSTAL STRUCTURE OF UREASE FROM KLEBSIELLA AEROGENES AT 2.2 ANGSTROMS RESOLUTION | | Descriptor: | NICKEL (II) ION, UREASE (ALPHA CHAIN), UREASE (BETA CHAIN), ... | | Authors: | Jabri, E, Carr, M.B, Hausinger, R.P, Karplus, P.A. | | Deposit date: | 1995-02-16 | | Release date: | 1995-07-10 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of urease from Klebsiella aerogenes.
Science, 268, 1995
|
|
3DK8
 
 | | Catalytic cycle of human glutathione reductase near 1 A resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE, GLYCEROL, ... | | Authors: | Berkholz, D.S, Faber, H.R, Savvides, S.N, Karplus, P.A. | | Deposit date: | 2008-06-24 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Catalytic cycle of human glutathione reductase near 1 A resolution.
J.Mol.Biol., 382, 2008
|
|
3LVB
 
 | | Crystal structure of the Ferredoxin:NADP+ reductase from maize root at 1.7 angstroms - Test Set Withheld | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin-NADP reductase | | Authors: | Faber, H.R, Karplus, P.A, Aliverti, A, Ferioli, C, Spinola, M. | | Deposit date: | 2010-02-19 | | Release date: | 2010-03-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and crystallographic characterization of ferredoxin-NADP(+) reductase from nonphotosynthetic tissues
Biochemistry, 40, 2001
|
|
3TF4
 
 | | ENDO/EXOCELLULASE:CELLOTRIOSE FROM THERMOMONOSPORA | | Descriptor: | CALCIUM ION, T. FUSCA ENDO/EXO-CELLULASE E4 CATALYTIC DOMAIN AND CELLULOSE-BINDING DOMAIN, beta-D-glucopyranose, ... | | Authors: | Sakon, J, Wilson, D.B, Karplus, P.A. | | Deposit date: | 1997-05-30 | | Release date: | 1997-09-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca.
Nat.Struct.Biol., 4, 1997
|
|
3E2B
 
 | | Crystal structure of Dynein Light chain LC8 in complex with a peptide derived from Swallow | | Descriptor: | ACETATE ION, Dynein light chain 1, cytoplasmic, ... | | Authors: | Benison, G, Barbar, E, Karplus, P.A. | | Deposit date: | 2008-08-05 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The interplay of ligand binding and quaternary structure in the diverse interactions of dynein light chain LC8.
J.Mol.Biol., 384, 2008
|
|
6B00
 
 | |
5TPR
 
 | |
5E0L
 
 | | LC8 - Chica (415-424) Complex | | Descriptor: | Dynein light chain 1, cytoplasmic, Protein Chica peptide, ... | | Authors: | Clark, S.A, Barbar, E.B, Karplus, P.A. | | Deposit date: | 2015-09-29 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | The Anchored Flexibility Model in LC8 Motif Recognition: Insights from the Chica Complex.
Biochemistry, 55, 2016
|
|
5E0M
 
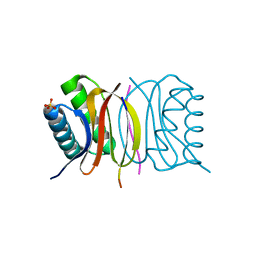 | | LC8 - Chica (468-476) Complex | | Descriptor: | Dynein light chain 1, cytoplasmic, Protein Chica peptide, ... | | Authors: | Clark, S.A, Barbar, E.B, Karplus, P.A. | | Deposit date: | 2015-09-29 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Anchored Flexibility Model in LC8 Motif Recognition: Insights from the Chica Complex.
Biochemistry, 55, 2016
|
|
6DVH
 
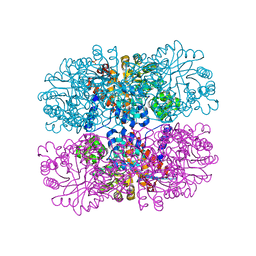 | |
6DVI
 
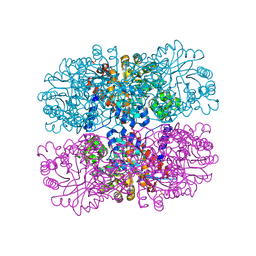 | |
8DQG
 
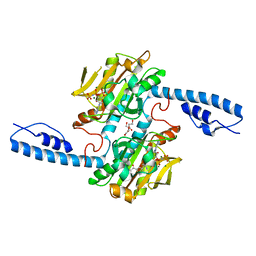 | | Crystal structure of pyrrolysyl-tRNA synthetase from Methanomethylophilus alvus engineered for acridone amino acid (RS1) bound to AMPPNP and acridone | | Descriptor: | (2~{S})-2-azanyl-3-(9-oxidanylidene-10~{H}-acridin-2-yl)propanoic acid, AA_TRNA_LIGASE_II domain-containing protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gottfried-Lee, I, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structures of Methanomethylophilus alvus Pyrrolysine tRNA-Synthetases Support the Need for De Novo Selections When Altering the Substrate Specificity.
Acs Chem.Biol., 17, 2022
|
|
8DQI
 
 | | Crystal structure of pyrrolysyl-tRNA synthetase from Methanomethylophilus alvus engineered for acridone amino acid (RS1) bound to ATP and acridone after 2- weeks of crystal growth | | Descriptor: | (2~{S})-2-azanyl-3-(9-oxidanylidene-10~{H}-acridin-2-yl)propanoic acid, AA_TRNA_LIGASE_II domain-containing protein, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gottfried-Lee, I, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structures of Methanomethylophilus alvus Pyrrolysine tRNA-Synthetases Support the Need for De Novo Selections When Altering the Substrate Specificity.
Acs Chem.Biol., 17, 2022
|
|
